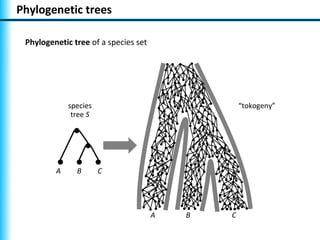
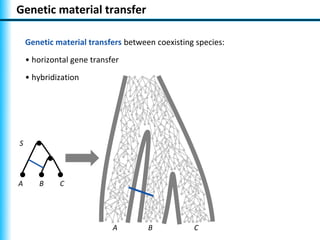
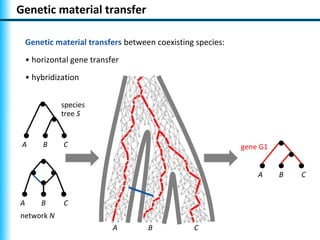
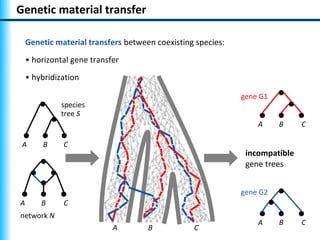
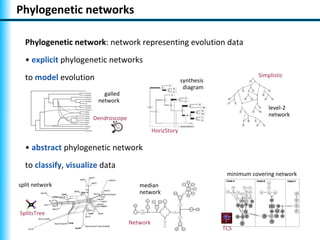
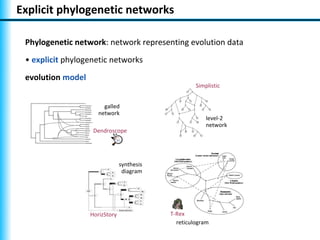
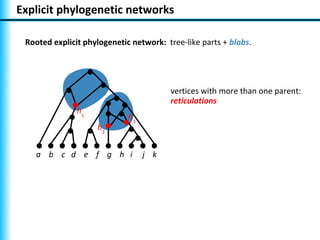
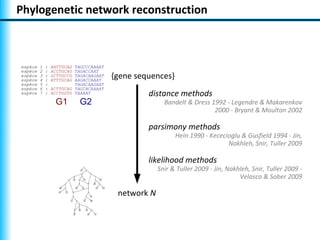
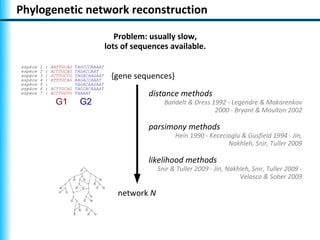
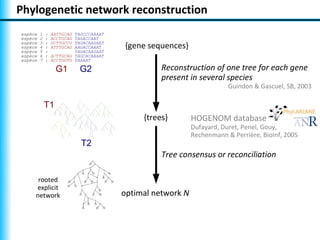
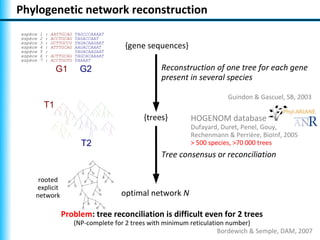
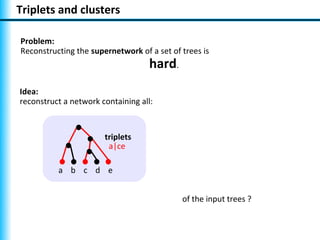
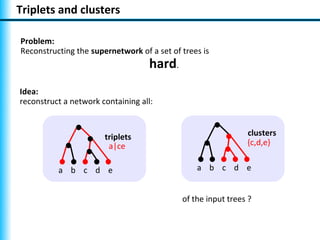
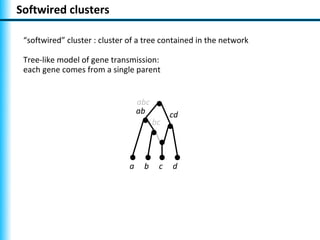
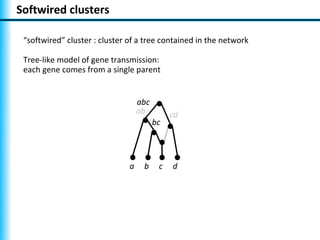
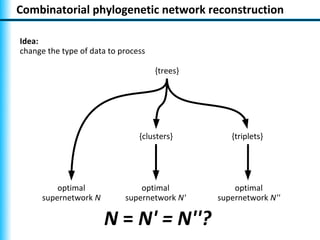
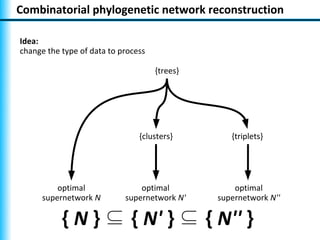
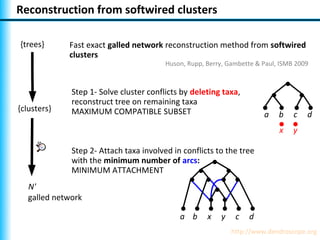
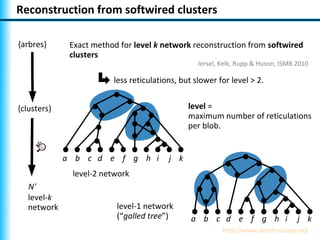
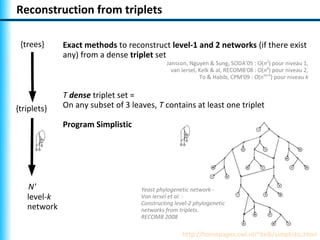
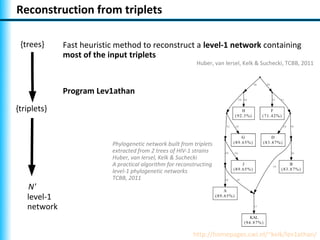
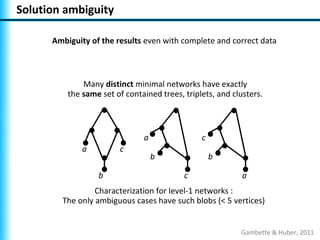
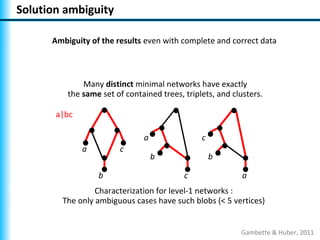
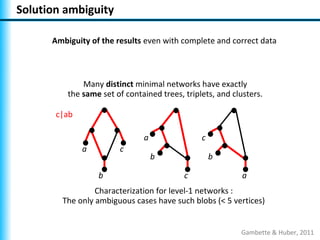
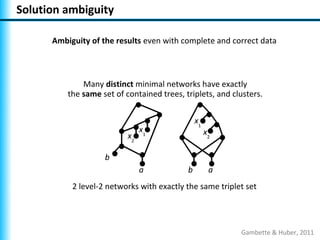
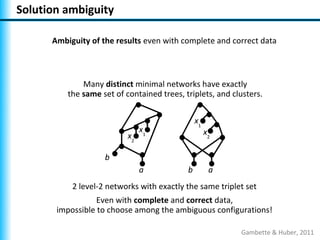
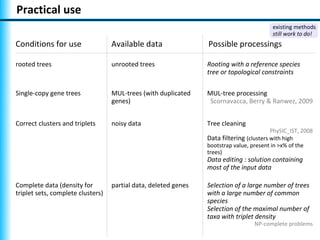
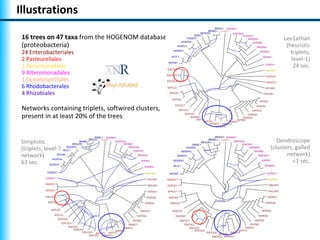
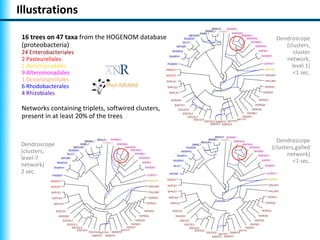
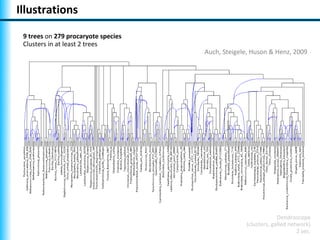
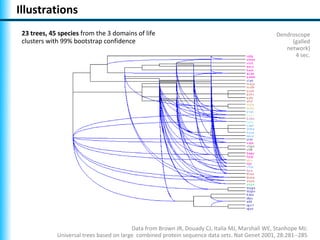
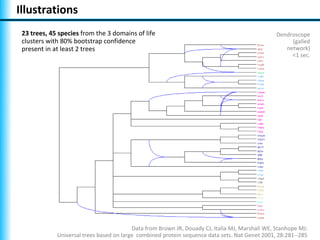
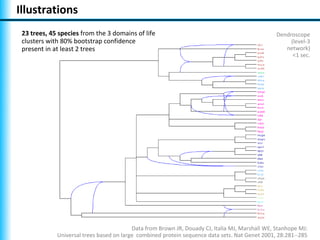
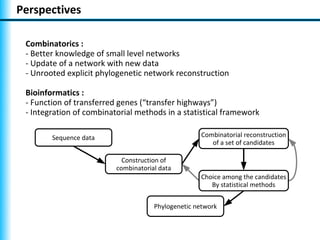
The document summarizes Philippe Gambette's presentation on the practical use of combinatorial methods for phylogenetic network reconstruction. It discusses phylogenetic networks as a way to model evolution when genetic material transfers occur between coexisting species. It motivates the combinatorial reconstruction approach of using clusters and triplets instead of whole trees as input data to make reconstruction faster. Combinatorial reconstruction methods are described that reconstruct networks from softwired clusters or triplets. The document illustrates applications on biological data and discusses challenges like solution ambiguity.