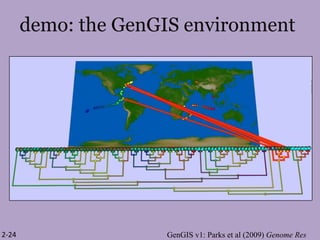
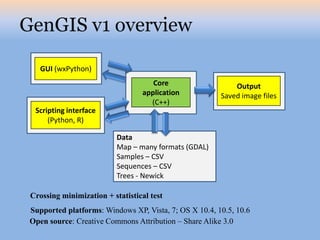
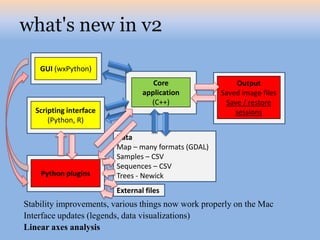
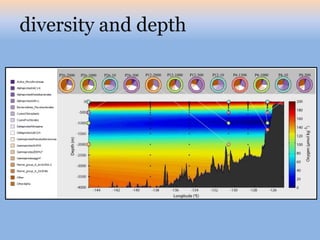
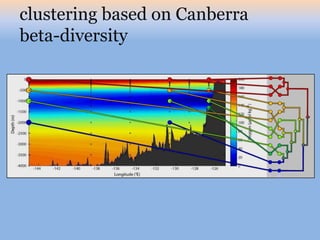
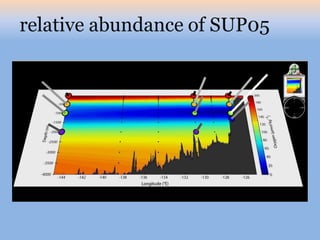
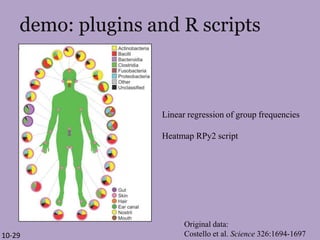
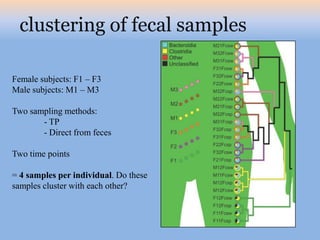
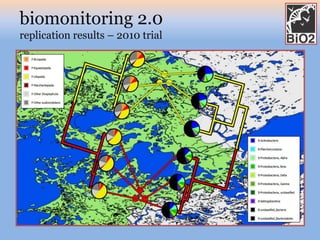
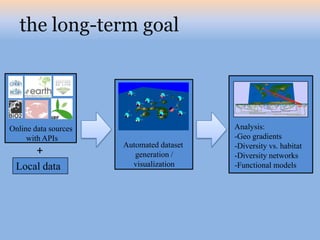
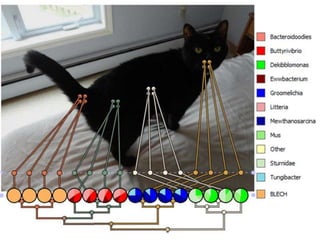
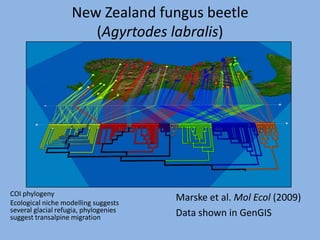
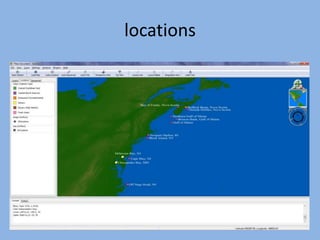
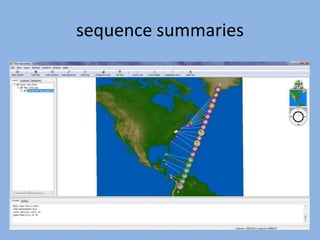
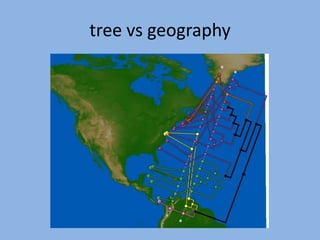
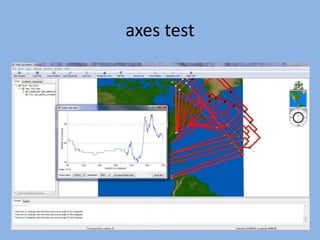
The document discusses the GENGIS software and its updated version, which enhances microbial world geography analysis through improved GUI, data handling, and plugins. It also outlines biomonitoring efforts in Wood Buffalo National Park, detailing sampling methods and objectives related to biodiversity assessment. The ongoing mission includes developing robust sampling techniques and inviting collaborators for future research initiatives.