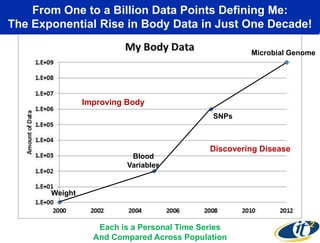
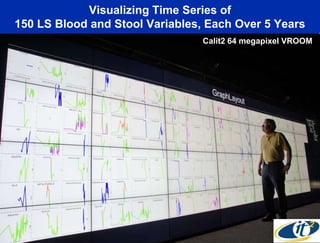
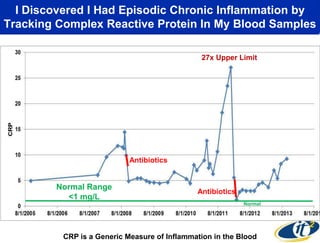
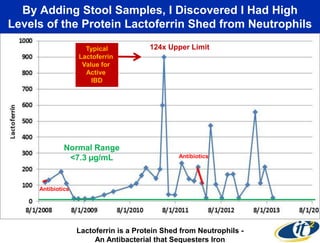
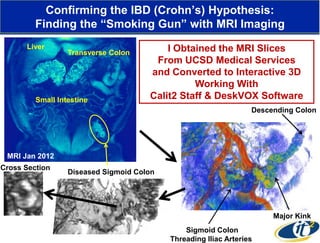
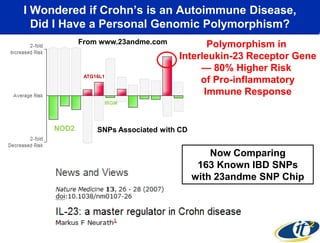
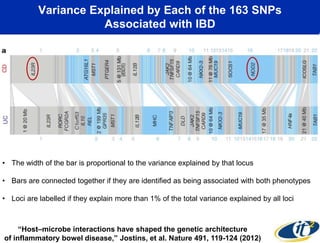
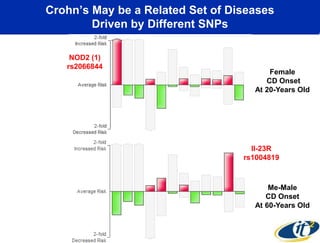
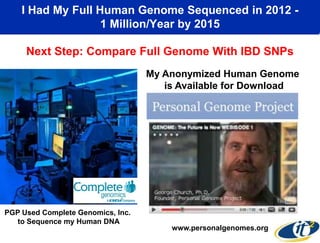
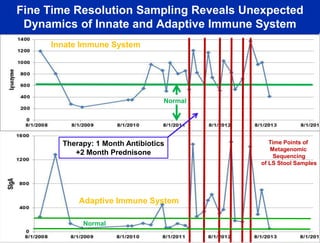
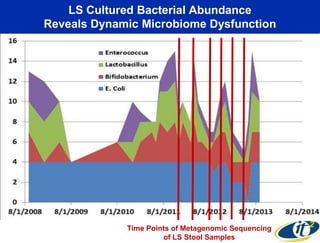
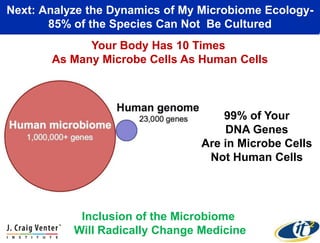
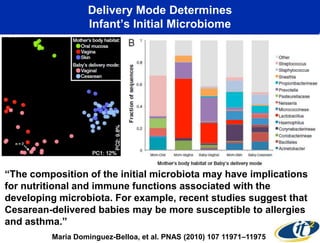
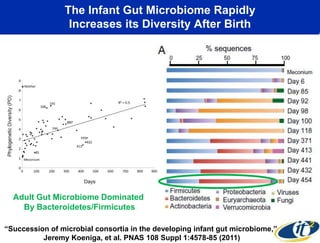
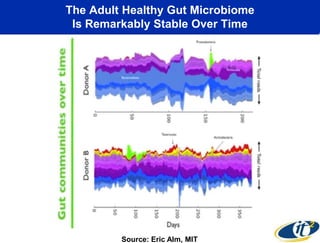
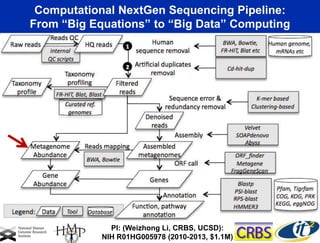
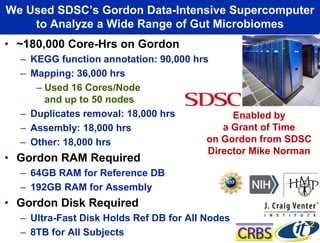
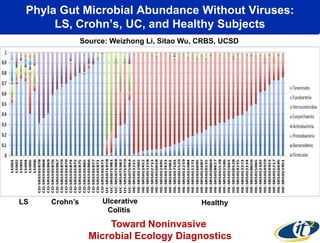
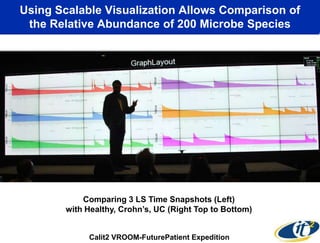
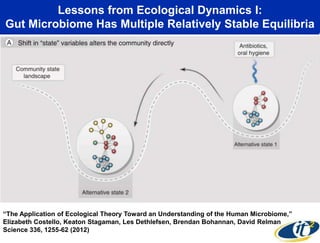
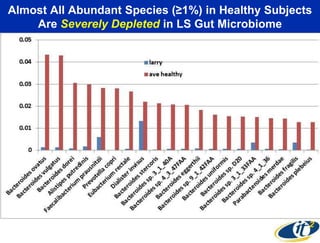
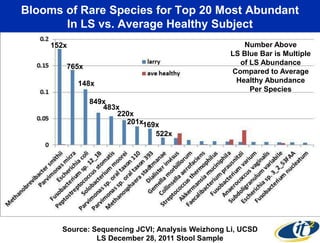
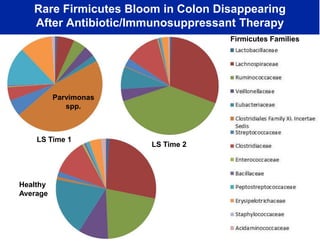
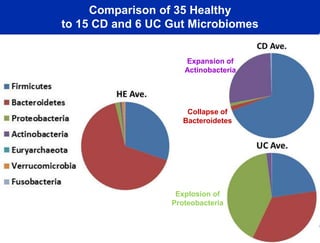
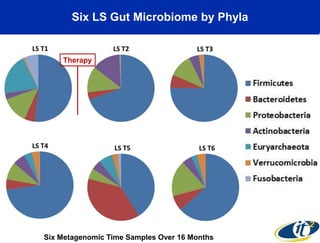
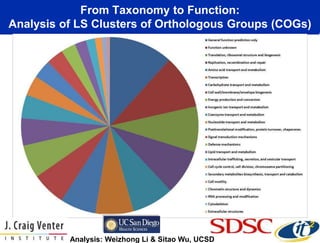
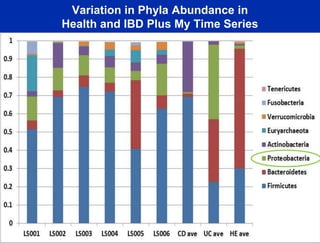
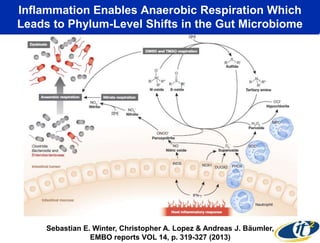
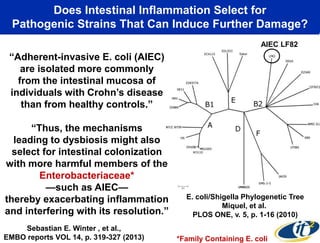
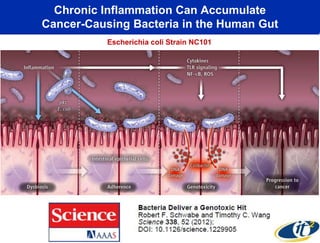
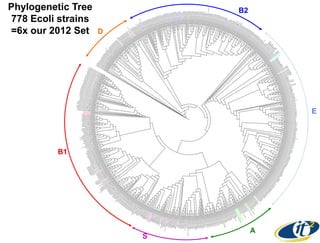
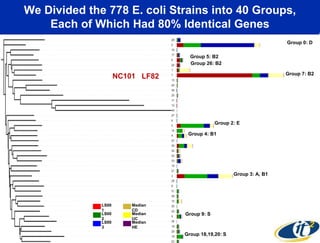
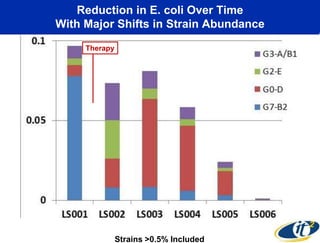
This document summarizes Dr. Larry Smarr's talk on modeling the human immune system and gut microbiome dynamics. It discusses how the growing diversity of gut bacteria after birth helps train the immune system. In health, constant feedback between the immune system and microbiome leads to homeostasis, but in diseases like Crohn's, this balance fails. The talk demonstrates this dysbiotic state using data from Dr. Smarr's own gut microbiome and biomarkers over five years. It reviews efforts to computationally model this important biological system.