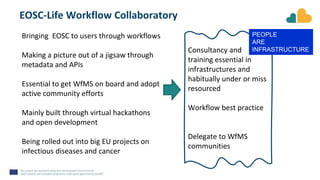
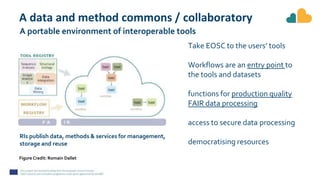
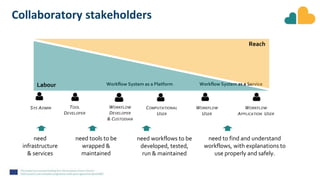
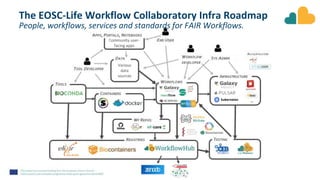
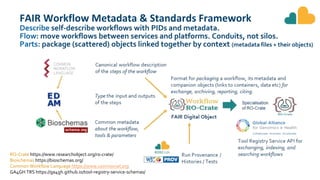
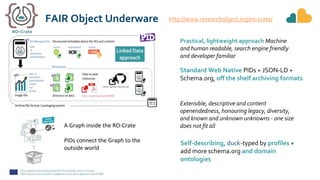
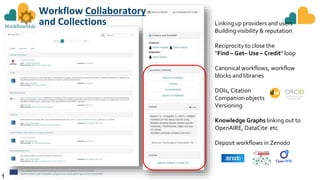
The document outlines the EOSC-Life Workflow Collaboratory, which aims to enhance access to and reuse of research data and tools within the European Open Science Cloud. It describes the collaborative environment supporting data-intensive bioscience research through sophisticated workflows, ensuring FAIR (Findable, Accessible, Interoperable, and Reusable) data practices. The initiative promotes interoperability between various workflow management systems and encourages community engagement towards the development and maintenance of workflows.



![Computational Workflows for Data intensive Bioscience
prepare, analyze, and share increasing volumes of complex data
CryoEM Image Analysis
Metagenomic Pipelines
Protein Ligand
Simulation
[Adam Hospital]
[Rob Finn]
[Carlos Oscar Sorzano Sanchez]
Nature 573, 149-150 (2019)
https://doi.org/10.1038/d41586-019-02619-z
Computational Workflows
Multi-step processes to
coordinate and execute
multiple codes and handle
data and processing
dependencies](https://image.slidesharecdn.com/eosc-life-wfcollaboratory-goble-211108114422/85/EOSC-Life-Workflow-Collaboratory-6-320.jpg)


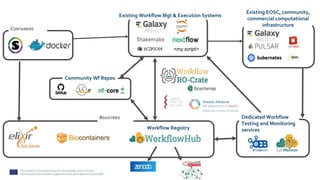


![Enable new services
https://github.com/inab/WfExS-backend
Workflow execution service for handling
sensitive human data & analysis
Consumes and creates RO-Crates
UI to start computational tasks based
on containerised software
[Jose Maria Fernandez, Laura Rodrigues-Navas, Salvador Capella, BSC]](https://image.slidesharecdn.com/eosc-life-wfcollaboratory-goble-211108114422/85/EOSC-Life-Workflow-Collaboratory-19-320.jpg)