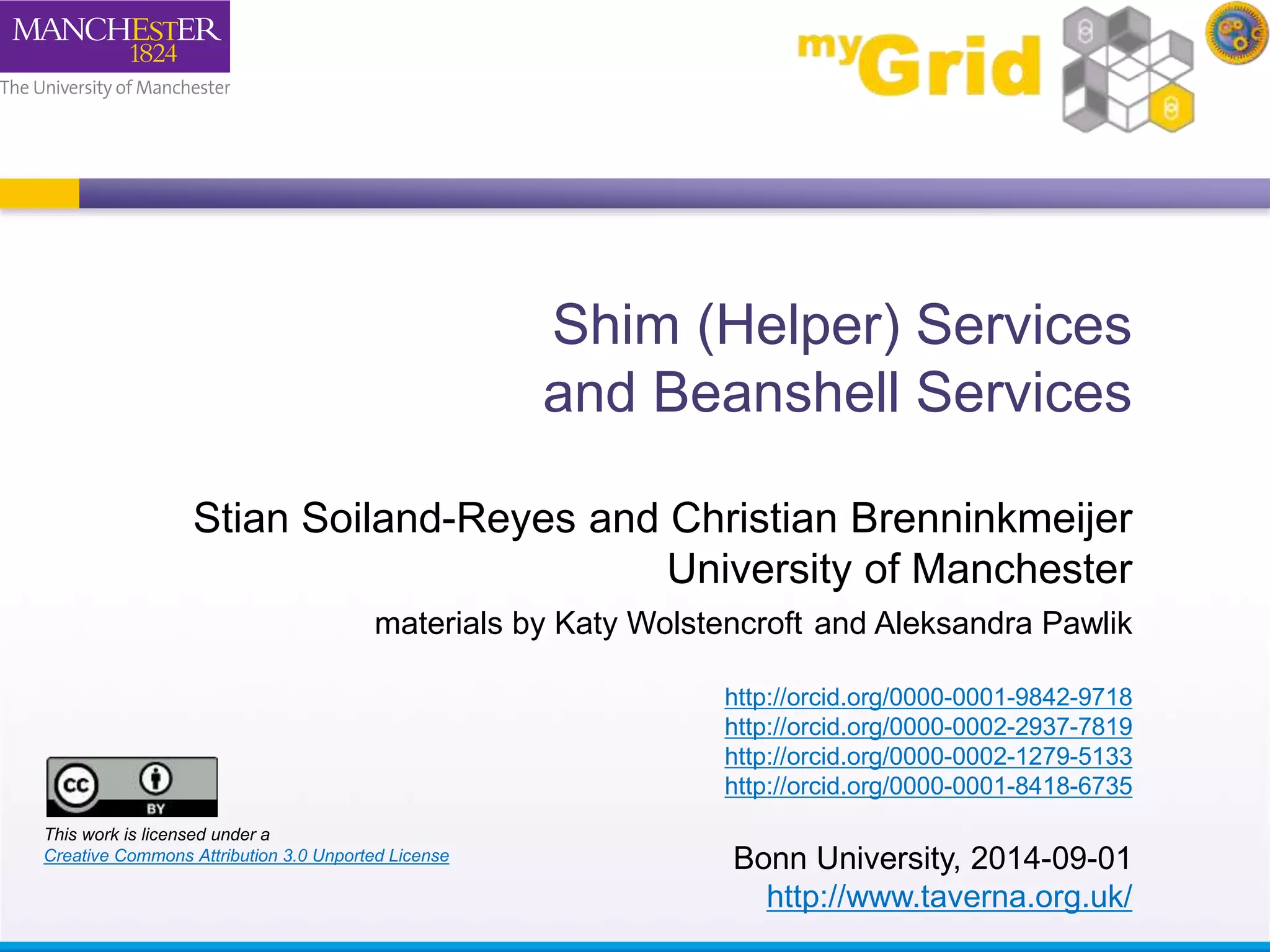
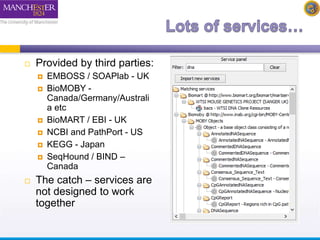
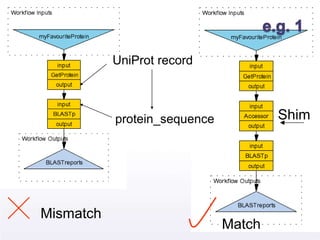
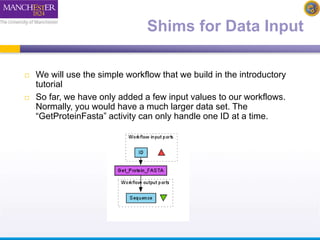
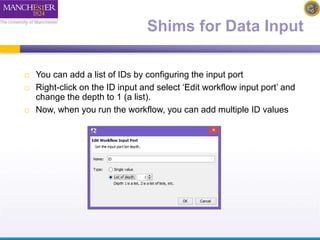
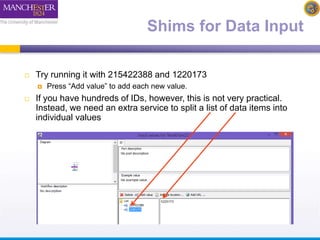
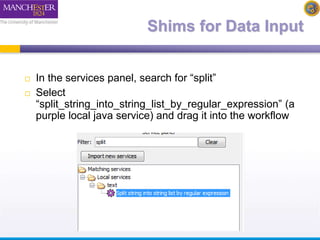
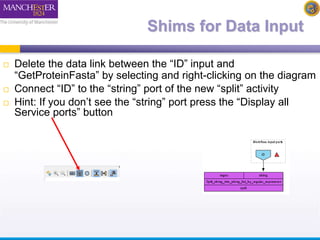
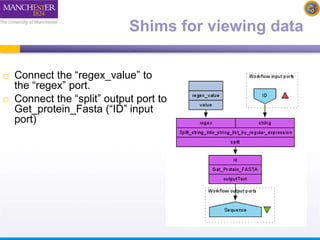
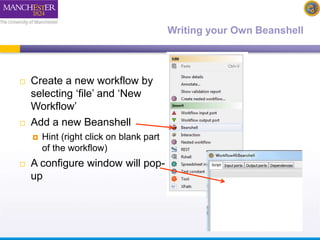
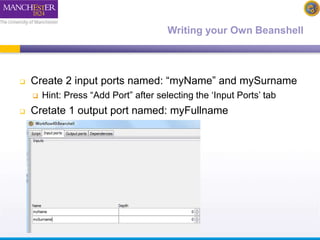
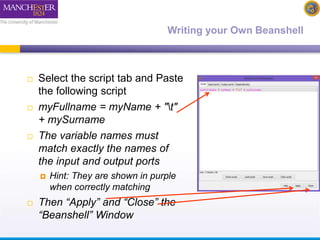
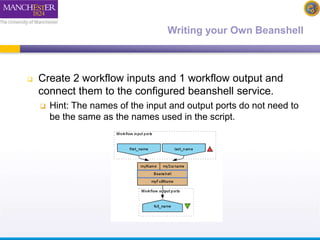
The document discusses shim and Beanshell services in Taverna workflows. It explains that shims act as connectors between incompatible scientific services, and are often simple Beanshell scripts. It provides examples of using split string and regex shims to handle multiple input values. The document also demonstrates how to write a simple Beanshell script to concatenate name and surname into a full name. The key points are that shims bridge incompatible services and Beanshells allow simple data transformations in workflows.