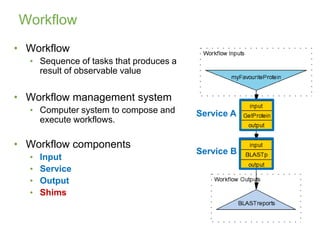
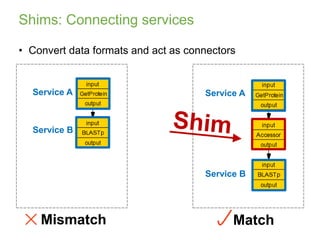
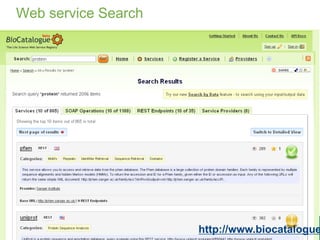
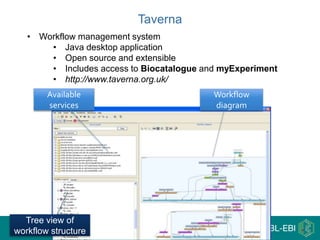
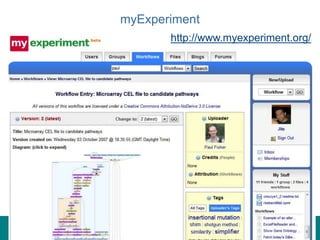
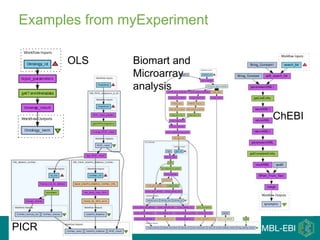
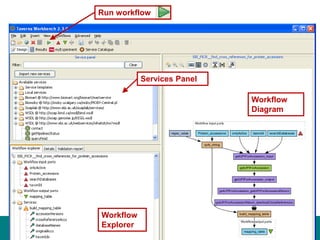
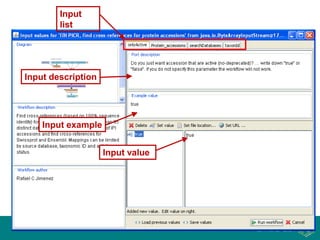
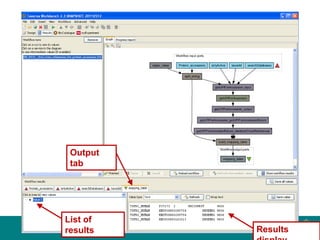
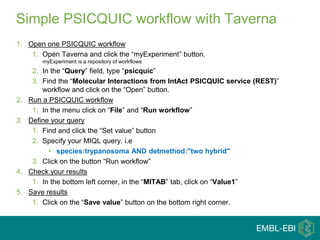
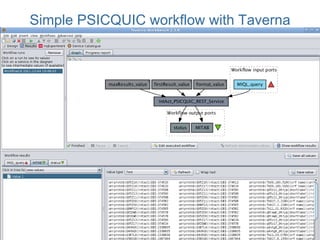
This document provides an introduction to workflows and web services at EBI. It discusses Taverna as a workflow management system that allows users to compose and execute workflows using various online biological databases and analysis tools as services. It also describes other myGrid solutions like Biocatalogue for discovering services and myExperiment for sharing workflows. Finally, it demonstrates a simple example of building a workflow in Taverna to retrieve molecular interactions from PSICQUIC services.