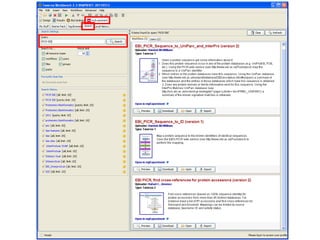
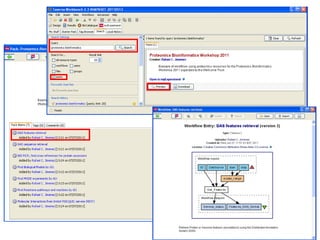
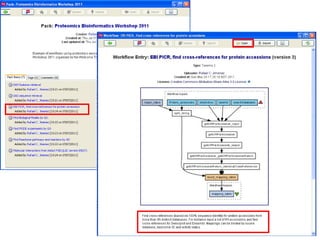
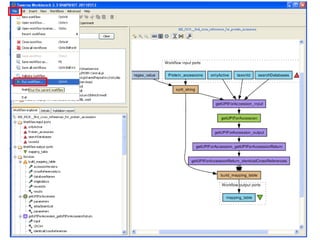
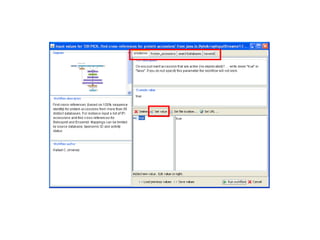
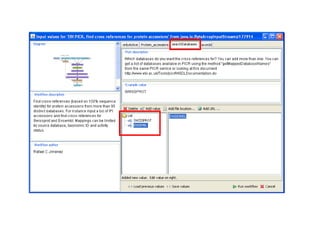
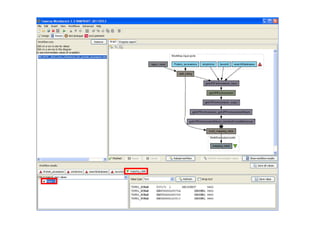
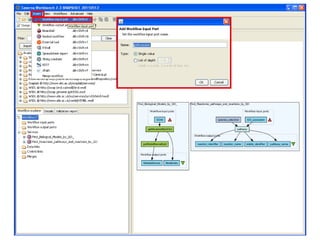
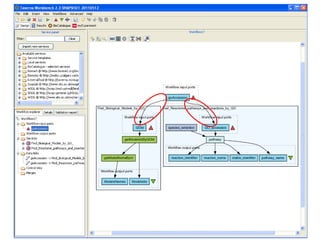
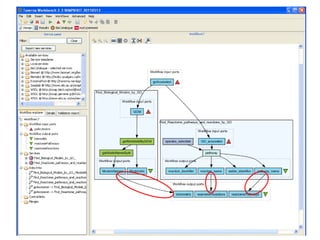
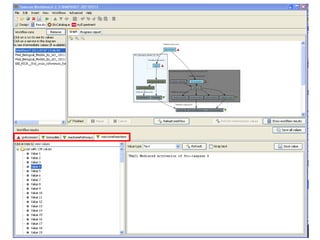
This document provides instructions for a series of exercises using the Taverna workflow management system. The exercises include searching for workflows on MyExperiment, running workflows to map protein accessions and perform BLAST searches, building a nested workflow, and running workflows from the command line. The goal is to familiarize users with searching, running, and constructing workflows in Taverna.