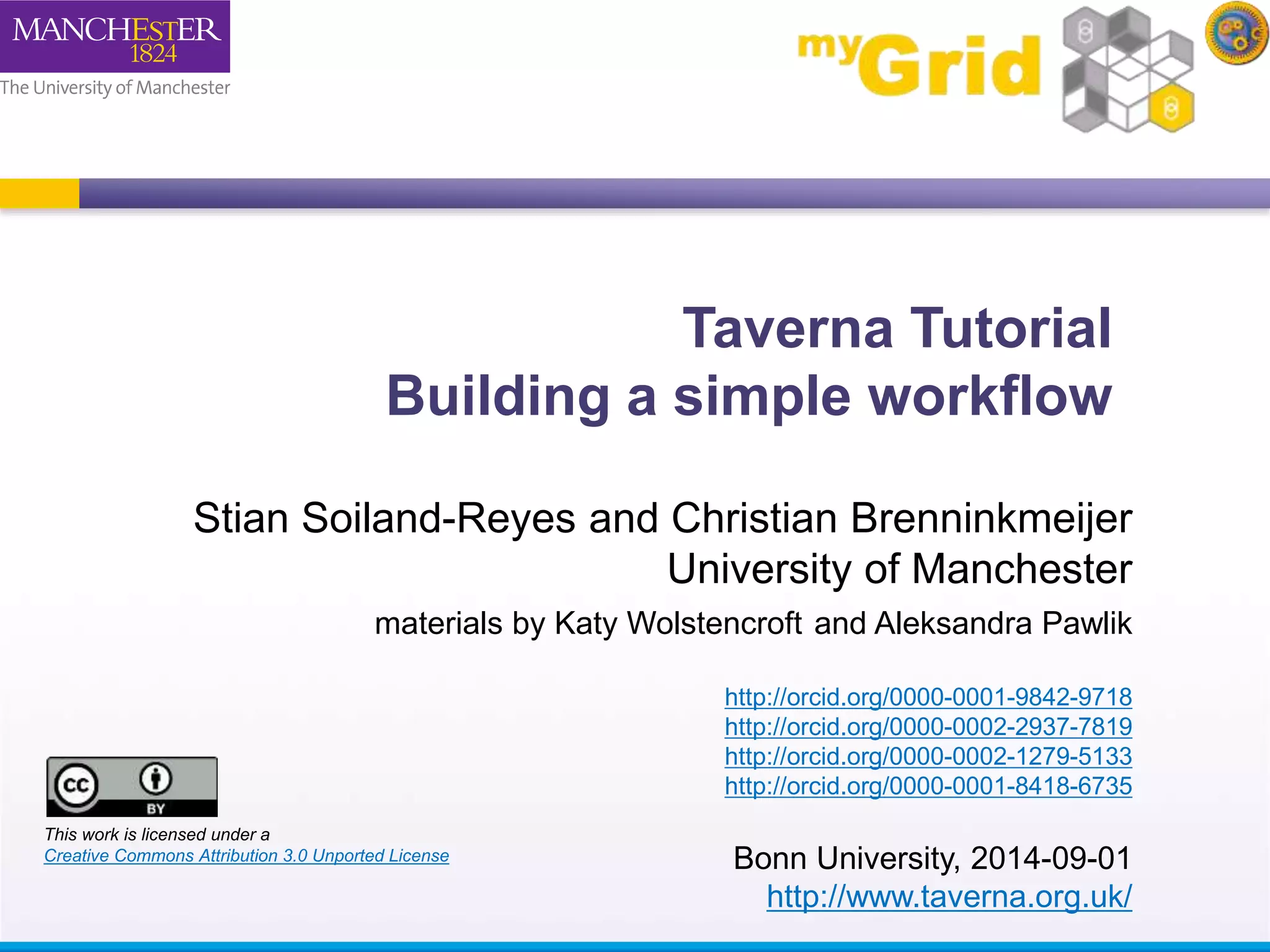
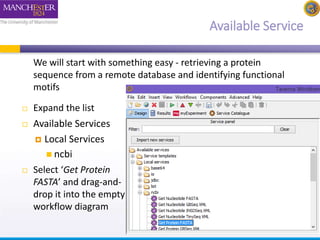
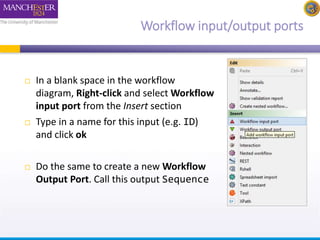
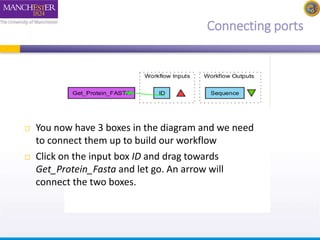
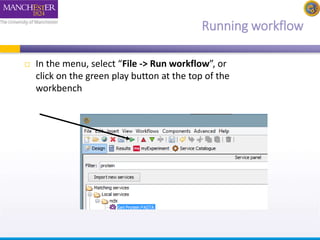
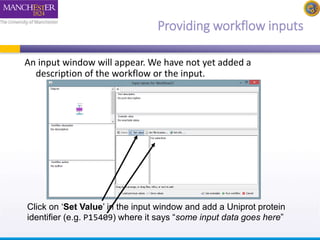
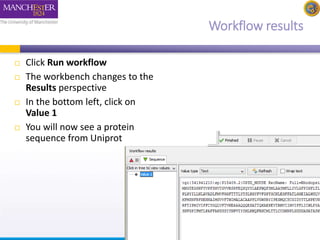
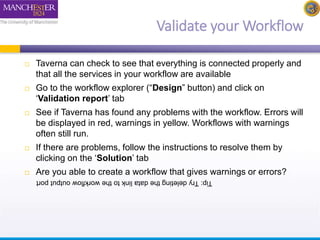
This document provides a tutorial on building a simple workflow in Taverna to retrieve a protein sequence from a remote database and identify functional motifs. It describes connecting input and output ports, running the workflow by providing a protein identifier, and viewing the resulting sequence output. It also explains how to validate that the workflow connections are proper and that the services are available.