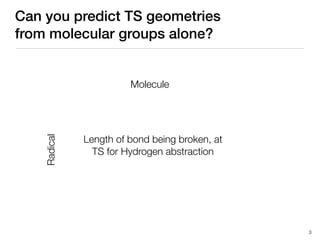
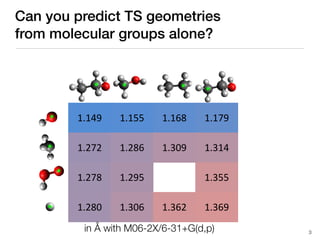
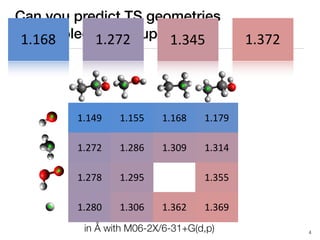
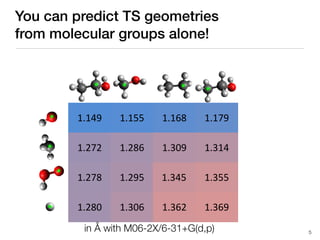
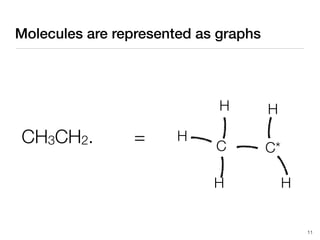
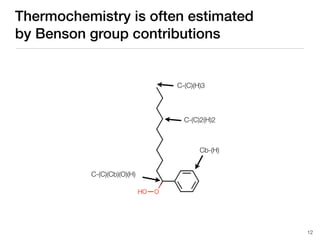
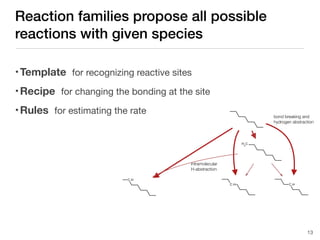
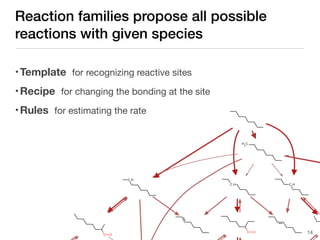
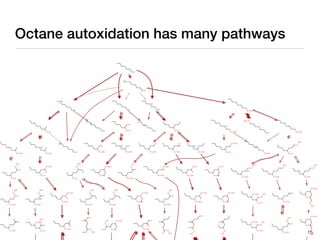
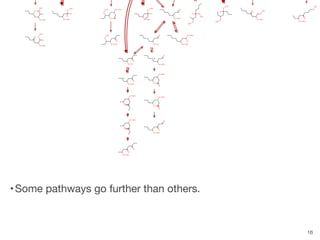
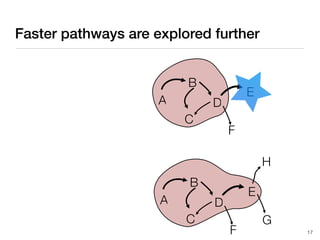
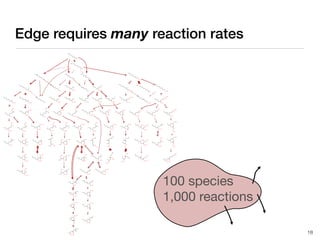
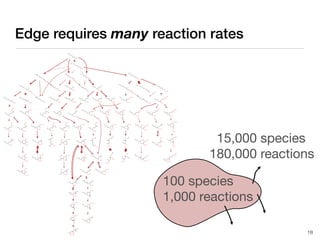
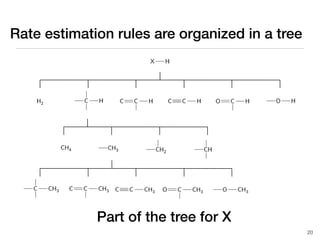
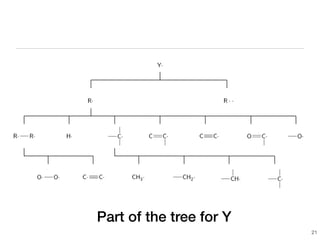
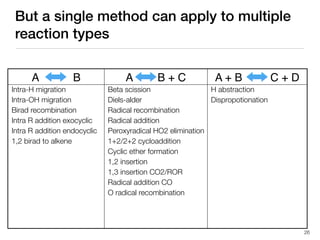
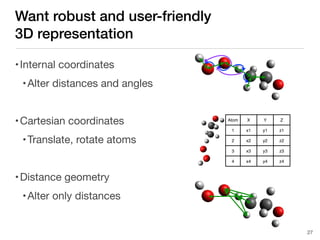
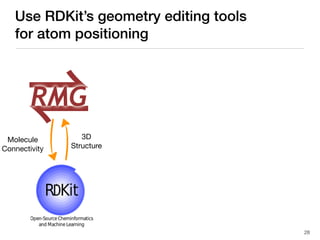
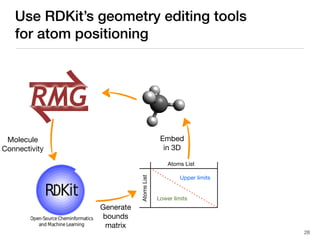
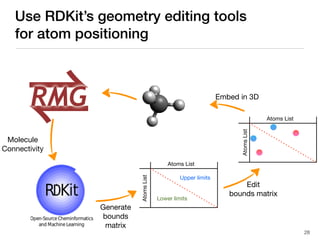
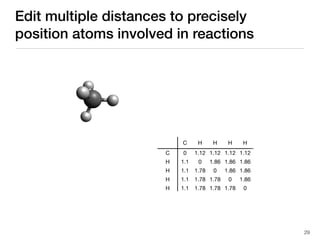
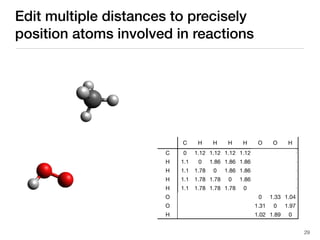
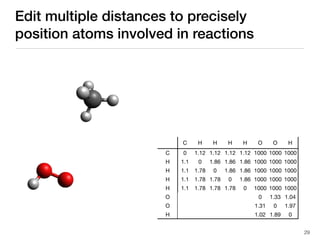
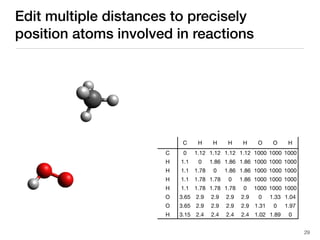
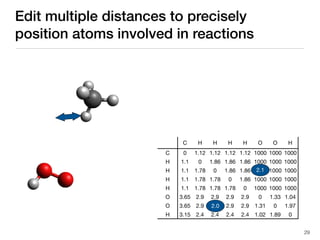
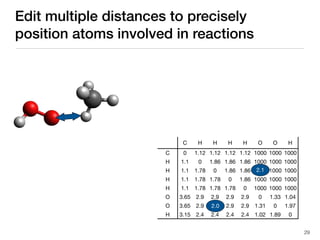
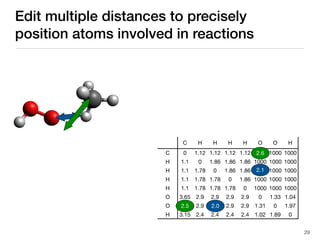
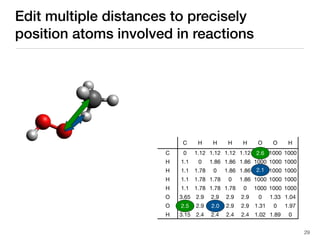
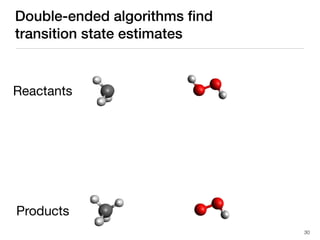
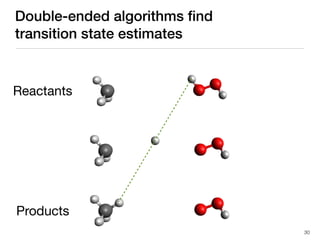
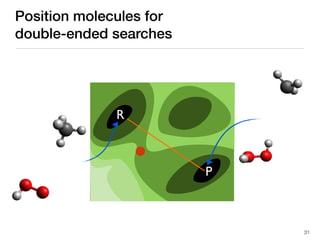
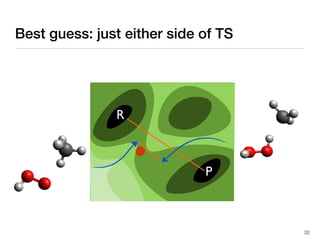
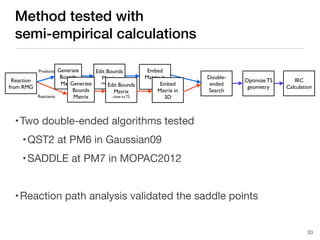
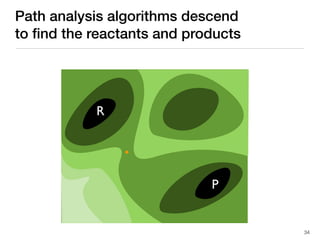
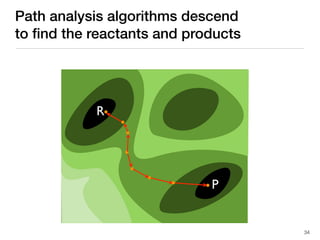
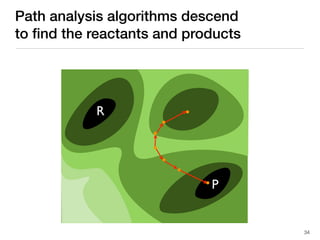
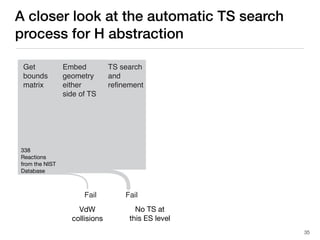
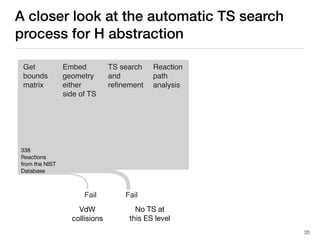
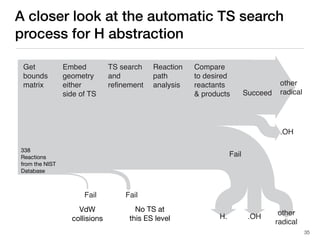
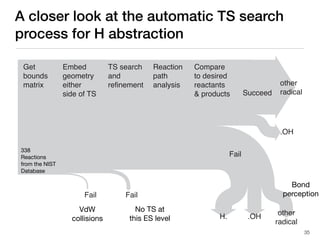
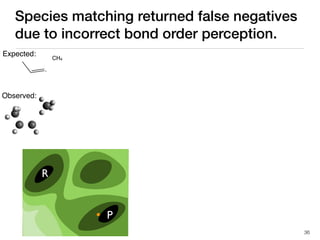
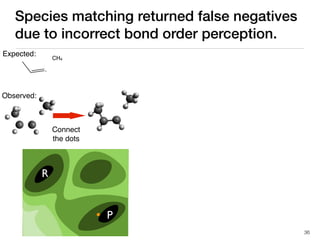
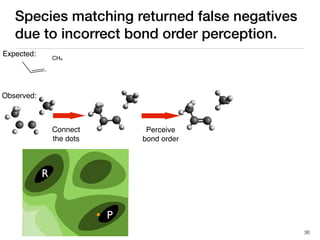
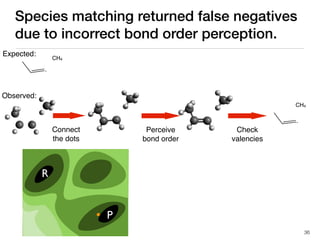
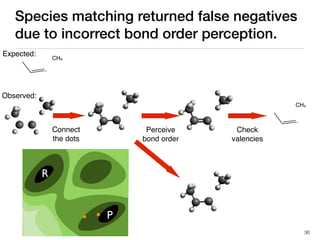
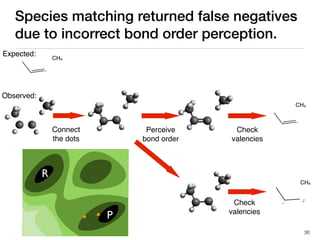
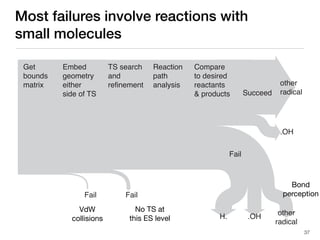
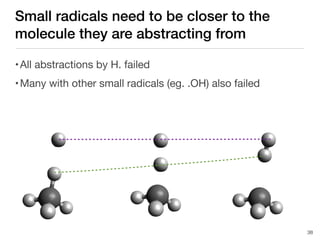
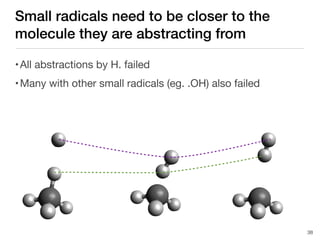
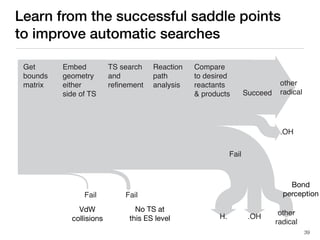
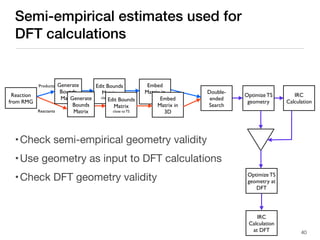
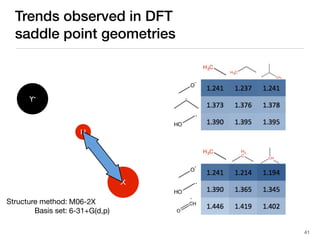
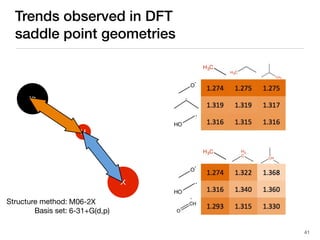
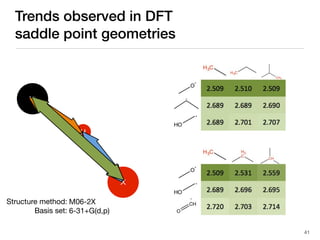
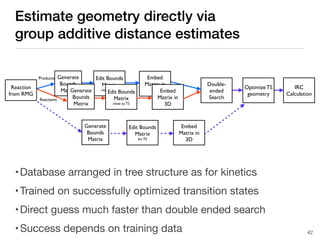
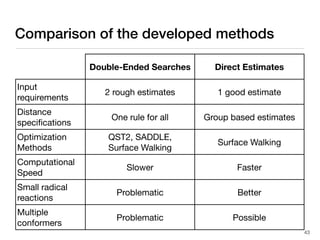
The document discusses advancements in computational modeling in chemical engineering, specifically focusing on the automatic generation of reaction mechanisms and transition state (TS) searches. It highlights the potential for predicting TS geometries from molecular groups and emphasizes the need for detailed kinetic models to enhance accuracy in energy research. The use of automated methods is portrayed as a transformative approach, enabling the characterization of complex reaction mechanisms and significantly speeding up calculations in the field of chemistry.