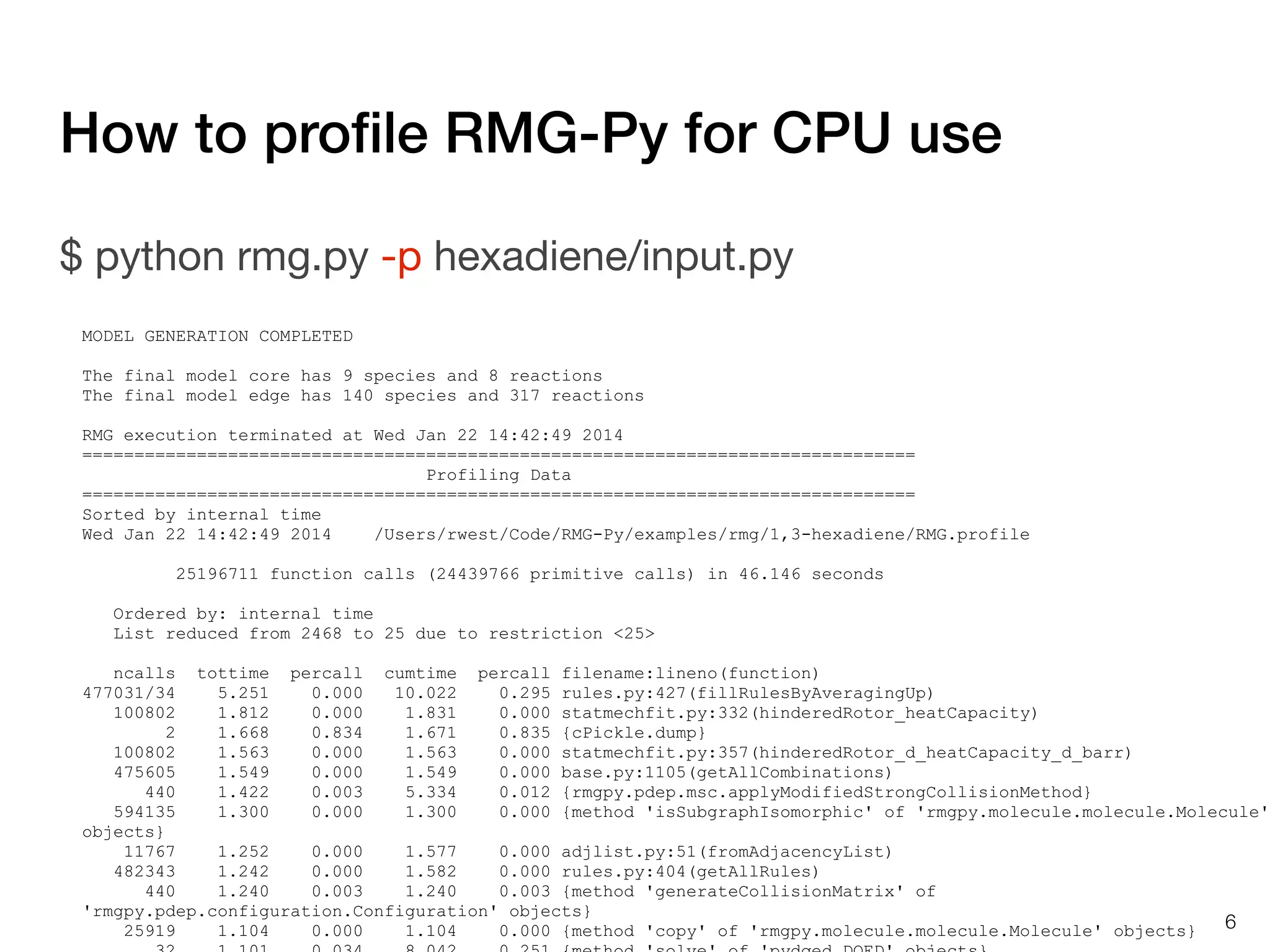
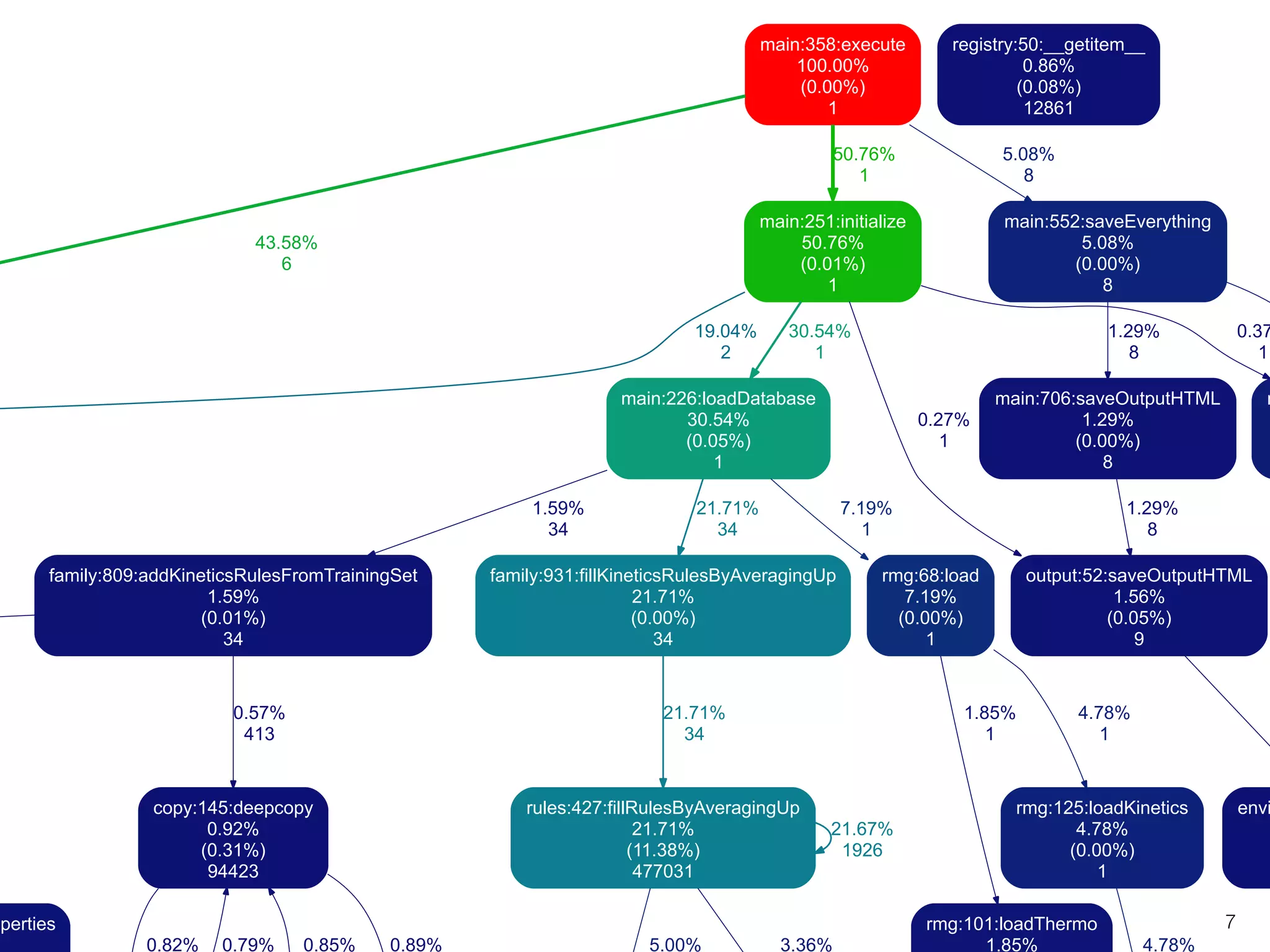
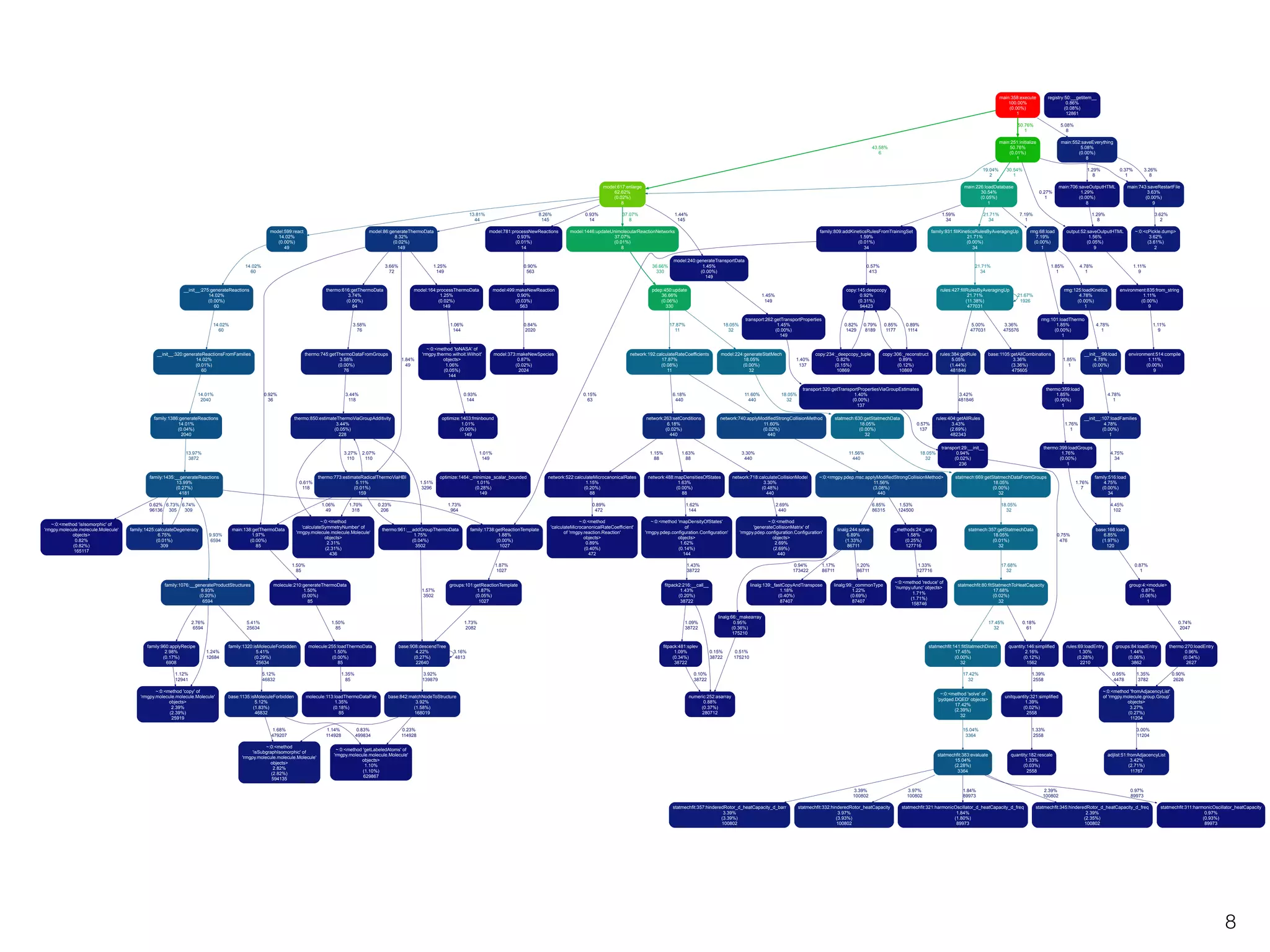
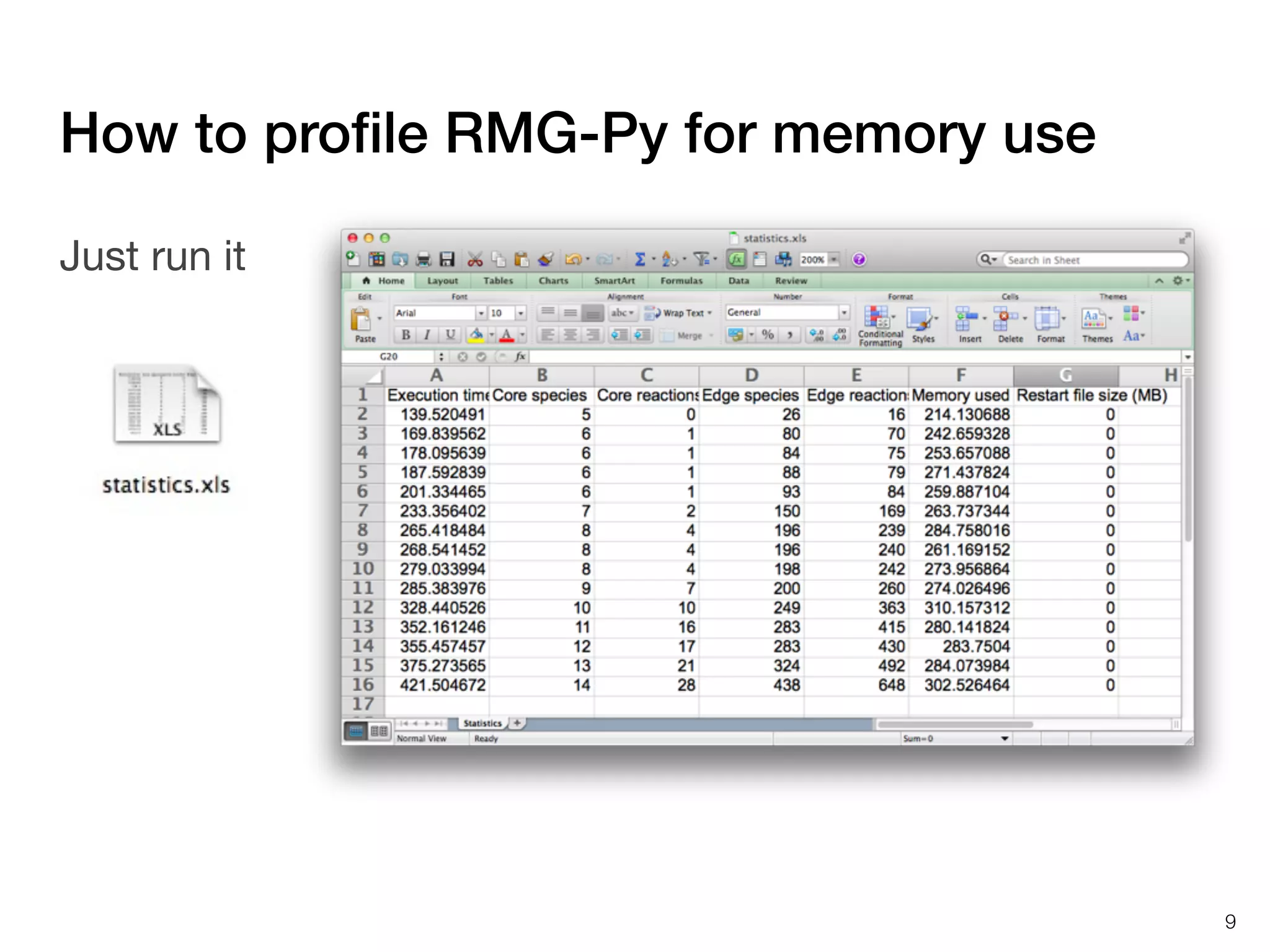
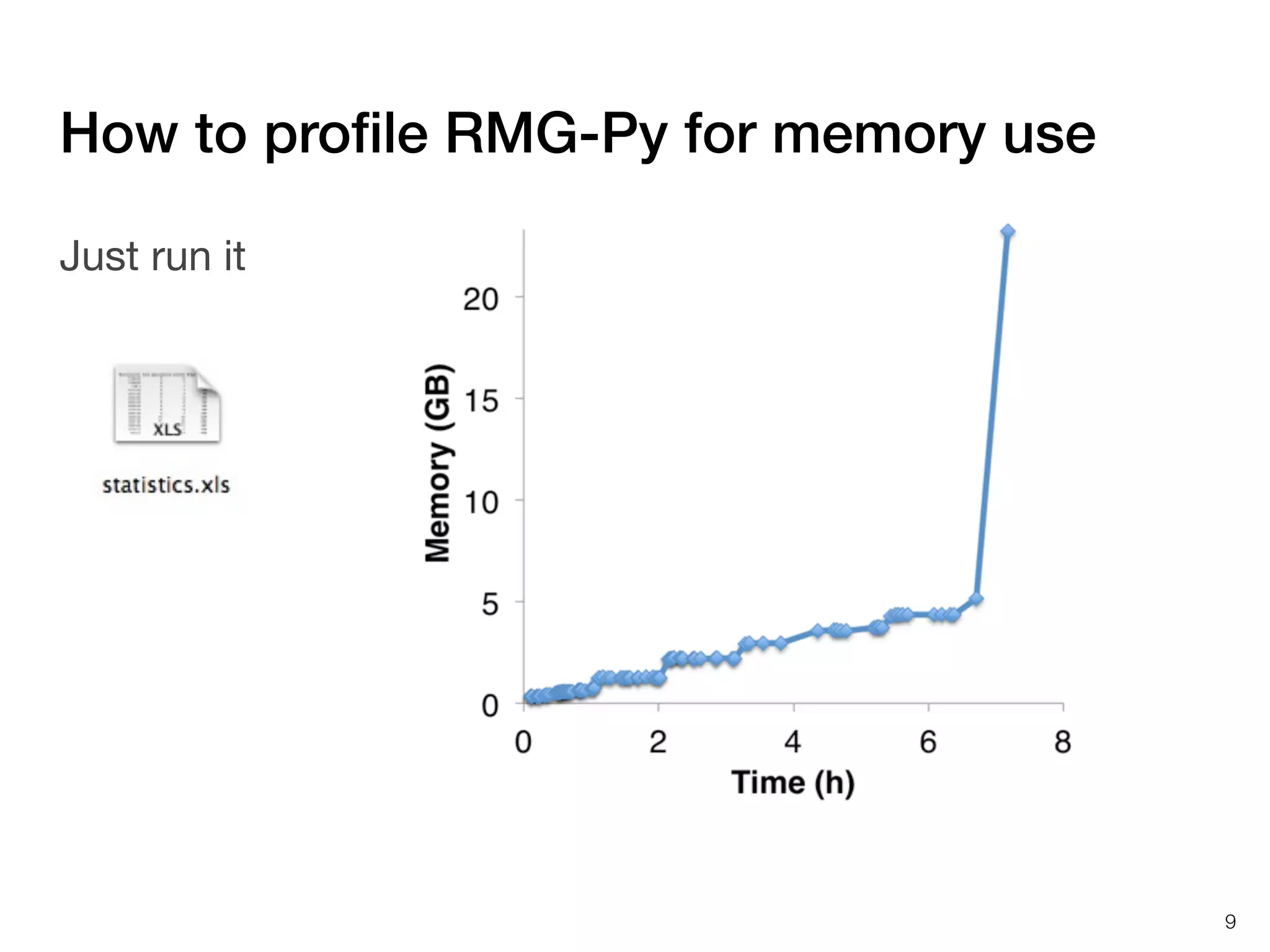
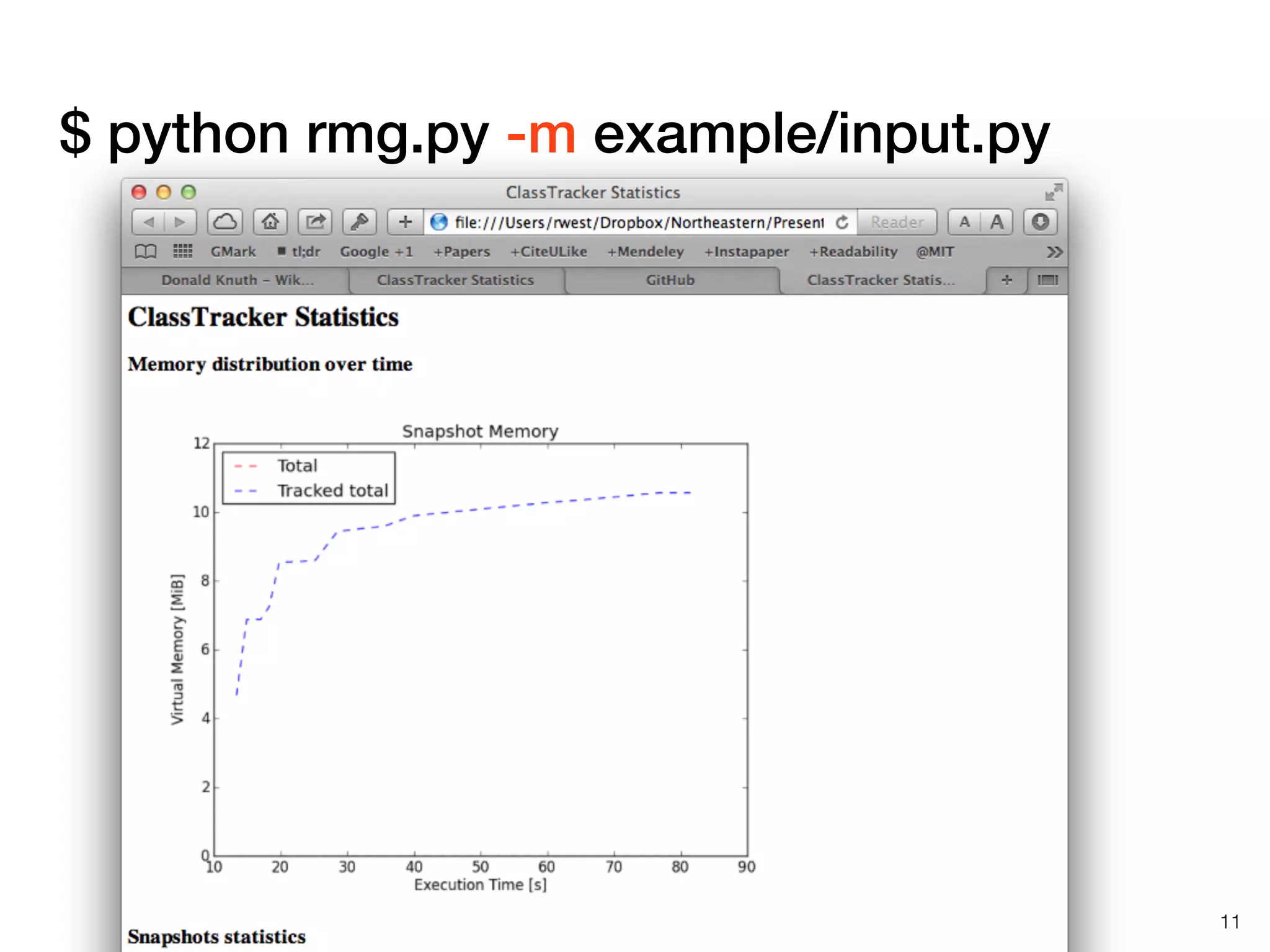
The document discusses profiling and optimizing RAM and CPU usage in RMG-Py. It begins with a quote about premature optimization being the root of evil. It then provides steps for profiling code, including getting it working, testing, profiling, and improving slow parts. The rest of the document shows profiling data from running RMG-Py on 1,3-hexadiene, with the most time spent on rules.fillRulesByAveragingUp and various statistical mechanics functions.