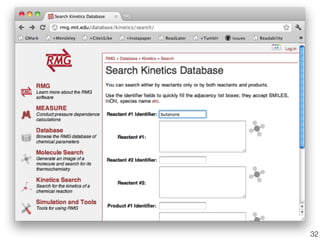
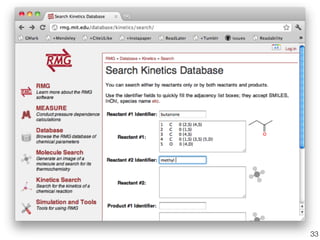
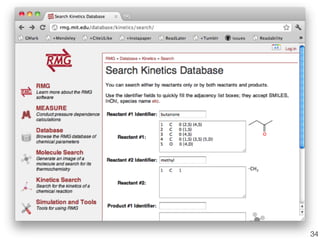
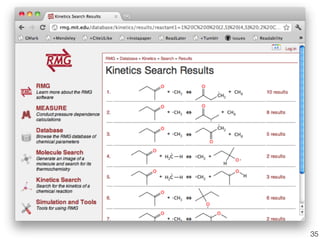
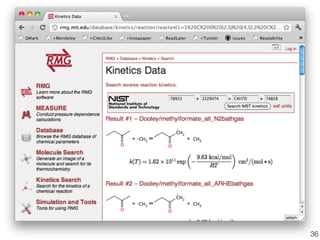
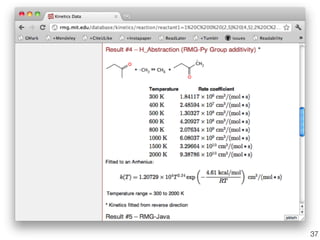
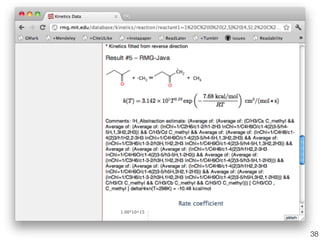
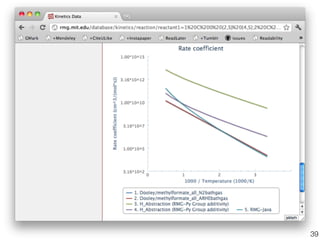
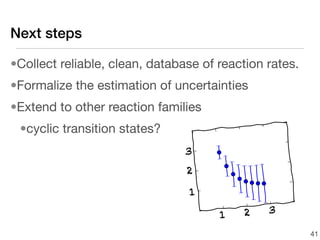
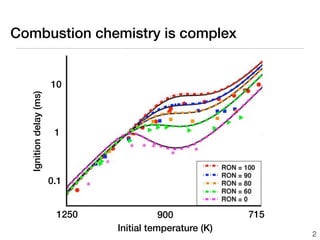
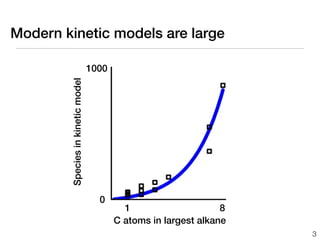
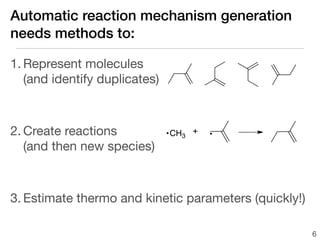
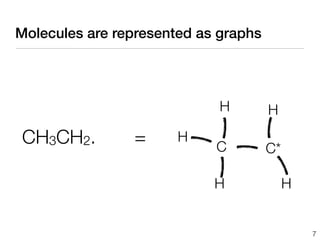
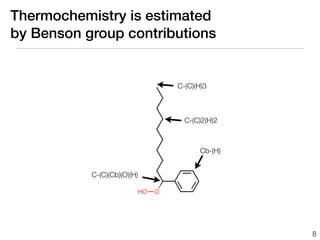
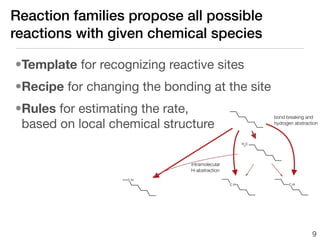
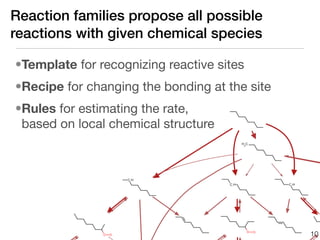
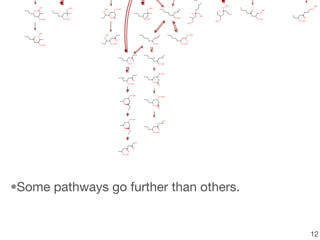
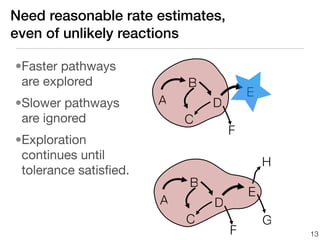
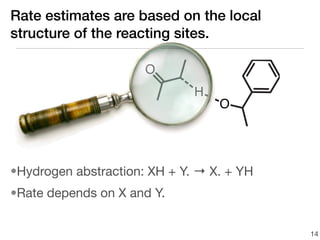
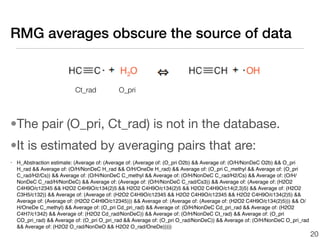
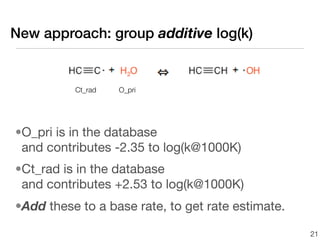
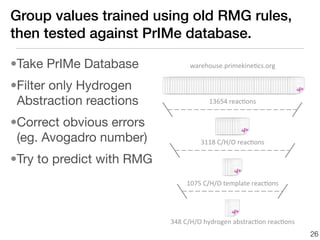
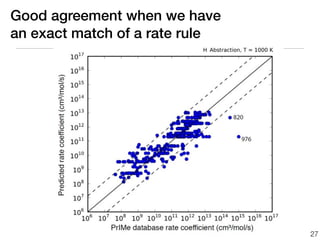
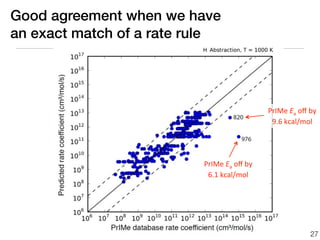
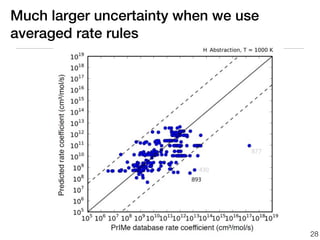
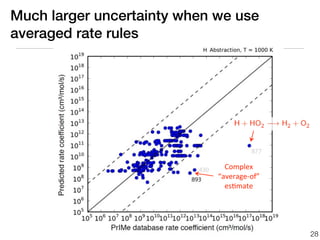
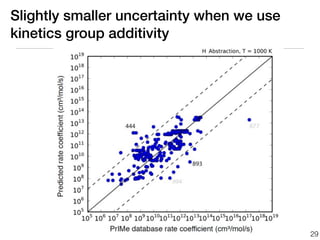
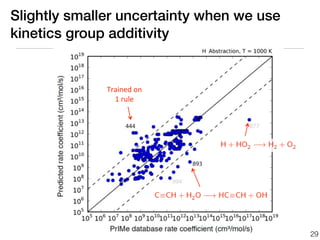
This document discusses using group additive kinetics to automatically generate chemical reaction mechanisms. It proposes representing molecules as graphs and using functional group contributions to estimate reaction rates. The approach is shown to work reasonably well for hydrogen abstraction reactions when the exact functional groups match the training data, but performs worse when using averaged rate rules. The document suggests using the group values to design a better reaction tree and collecting more reliable kinetic data to further improve the method.

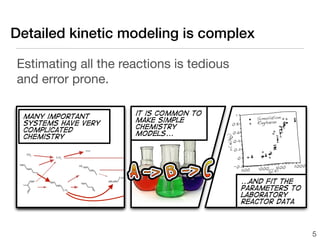
![Detailed kinetic modeling is complex
For each chemical reaction
A + B C + D we need:
•forward rate coefficient
r = k f [A][B]
− Ea
k f = A exp
•equilibrium constant RT
kf −∆G
= Keq = exp
kr RT
∆G = ∆H − T∆S 4](https://image.slidesharecdn.com/aichetalk-rmgwithgroupadditivekinetics-111023200051-phpapp01/85/AIChE-2011-Automatic-Reaction-Mechanism-Generation-with-Group-Additive-Kinetics-4-320.jpg)



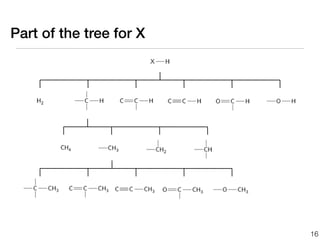





![We can use the group values
to design a better tree
log
kXH(1000
K)
[cm3/mol*s] ±0.0
(233) Number
of
entries
trained
against
30](https://image.slidesharecdn.com/aichetalk-rmgwithgroupadditivekinetics-111023200051-phpapp01/85/AIChE-2011-Automatic-Reaction-Mechanism-Generation-with-Group-Additive-Kinetics-36-320.jpg)
![We can use the group values
to design a better tree
log
kXH(1000
K)
[cm3/mol*s] ±0.0
(233) Number
of
entries
trained
against
-‐0.59
(19) +0.18
(120) -‐0.55
(25) -‐2.31
(5) +0.79
(22) -‐0.05
(34)
-‐0.45
(16) +0.16
(47) +0.18
(28) +0.56
(29)
-‐0.12
(17) +0.05
(23) +1.83
(2) +2.09
(1) +1.42
(3)
30](https://image.slidesharecdn.com/aichetalk-rmgwithgroupadditivekinetics-111023200051-phpapp01/85/AIChE-2011-Automatic-Reaction-Mechanism-Generation-with-Group-Additive-Kinetics-37-320.jpg)
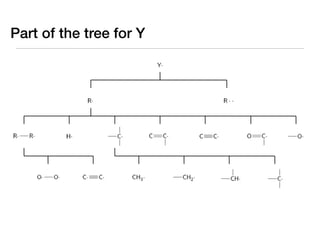
![We can use the group values
to design a better tree
log
kY.(1000
K)
[cm3/mol*s] ±0.0
(233) Number
of
entries
trained
against
31](https://image.slidesharecdn.com/aichetalk-rmgwithgroupadditivekinetics-111023200051-phpapp01/85/AIChE-2011-Automatic-Reaction-Mechanism-Generation-with-Group-Additive-Kinetics-38-320.jpg)
![We can use the group values
to design a better tree
log
kY.(1000
K)
[cm3/mol*s] ±0.0
(233) Number
of
entries
trained
against
-‐0.09
(218) +1.68
(13)
-‐7.01
(13) +2.21
(23) -‐0.56
(97) +1.02
(26) +2.62
(7) -‐1.13
(12) +0.77
(37)
-‐7.82
(12) +3.52
(1) +0.54
(23) -‐0.69
(34) -‐0.96
(23) -‐1.11
(17)
31](https://image.slidesharecdn.com/aichetalk-rmgwithgroupadditivekinetics-111023200051-phpapp01/85/AIChE-2011-Automatic-Reaction-Mechanism-Generation-with-Group-Additive-Kinetics-39-320.jpg)