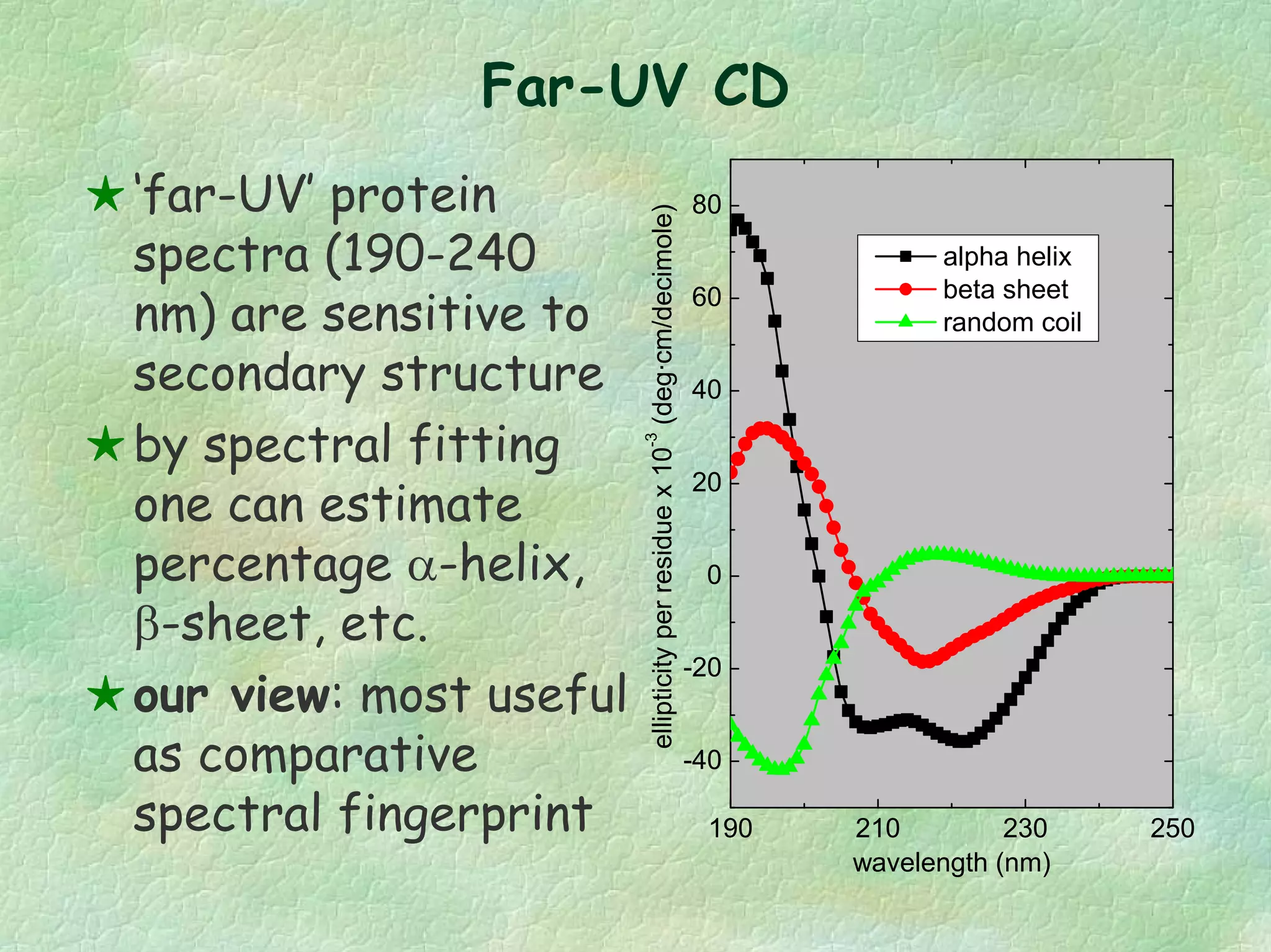
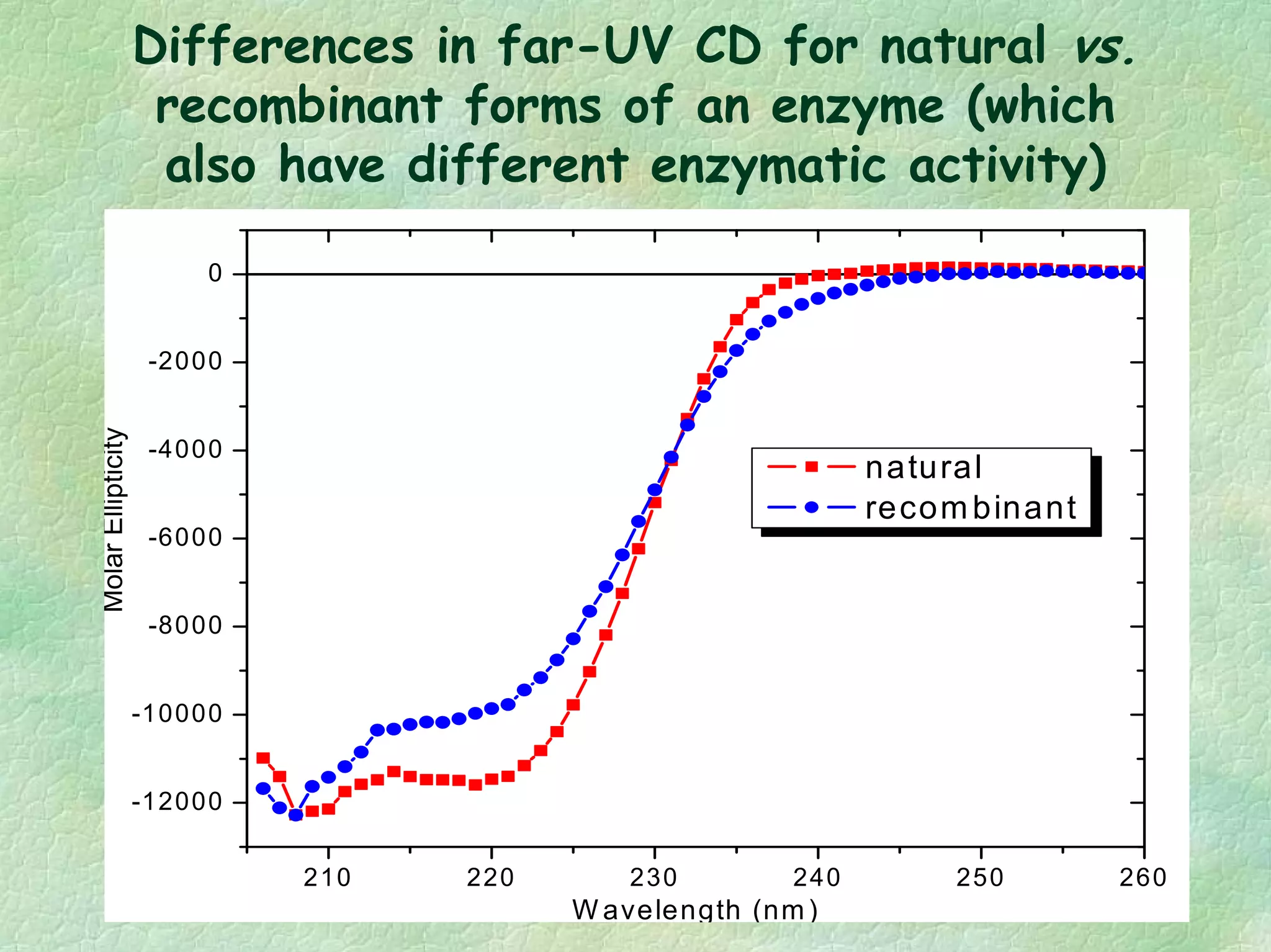
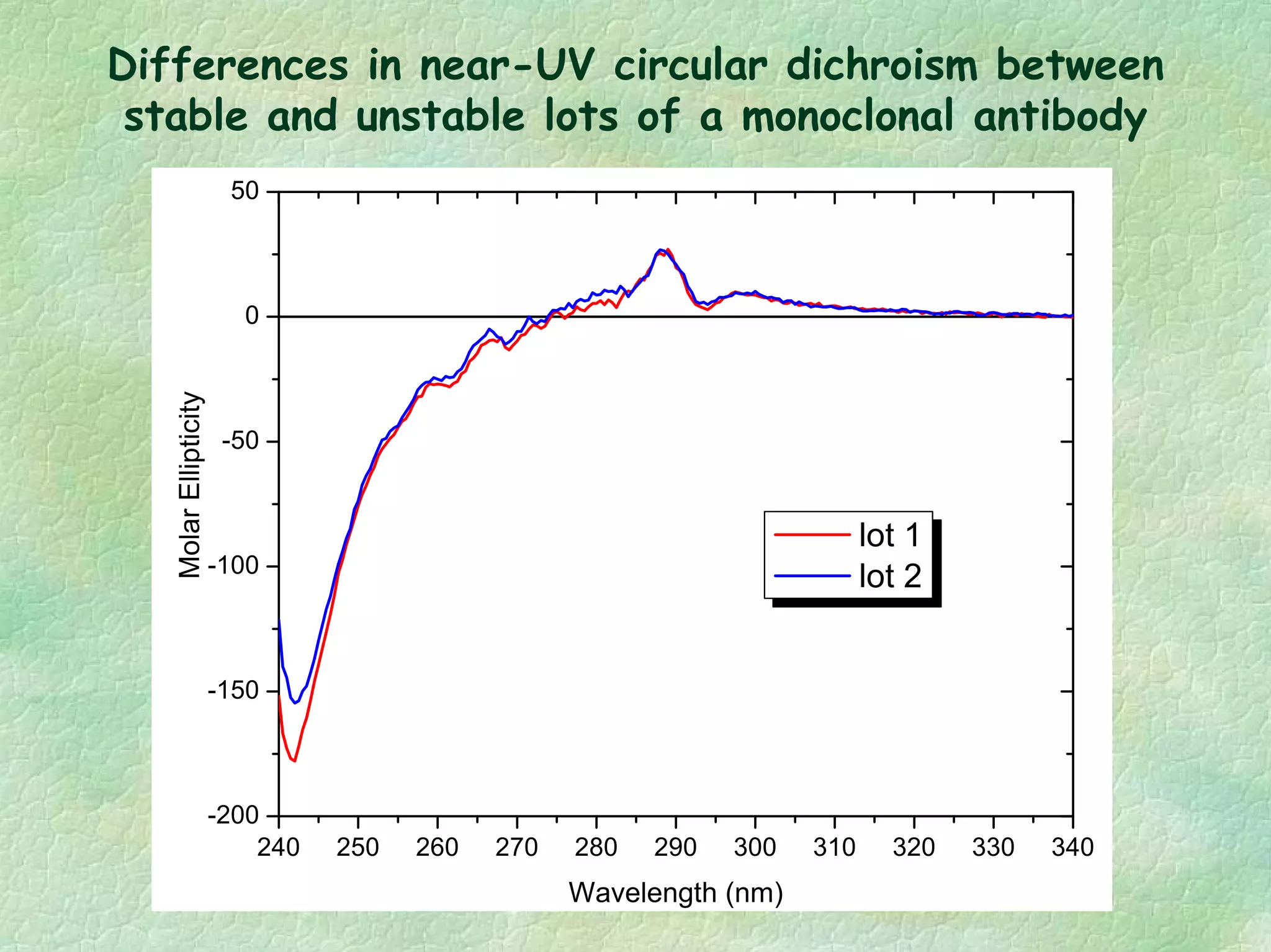
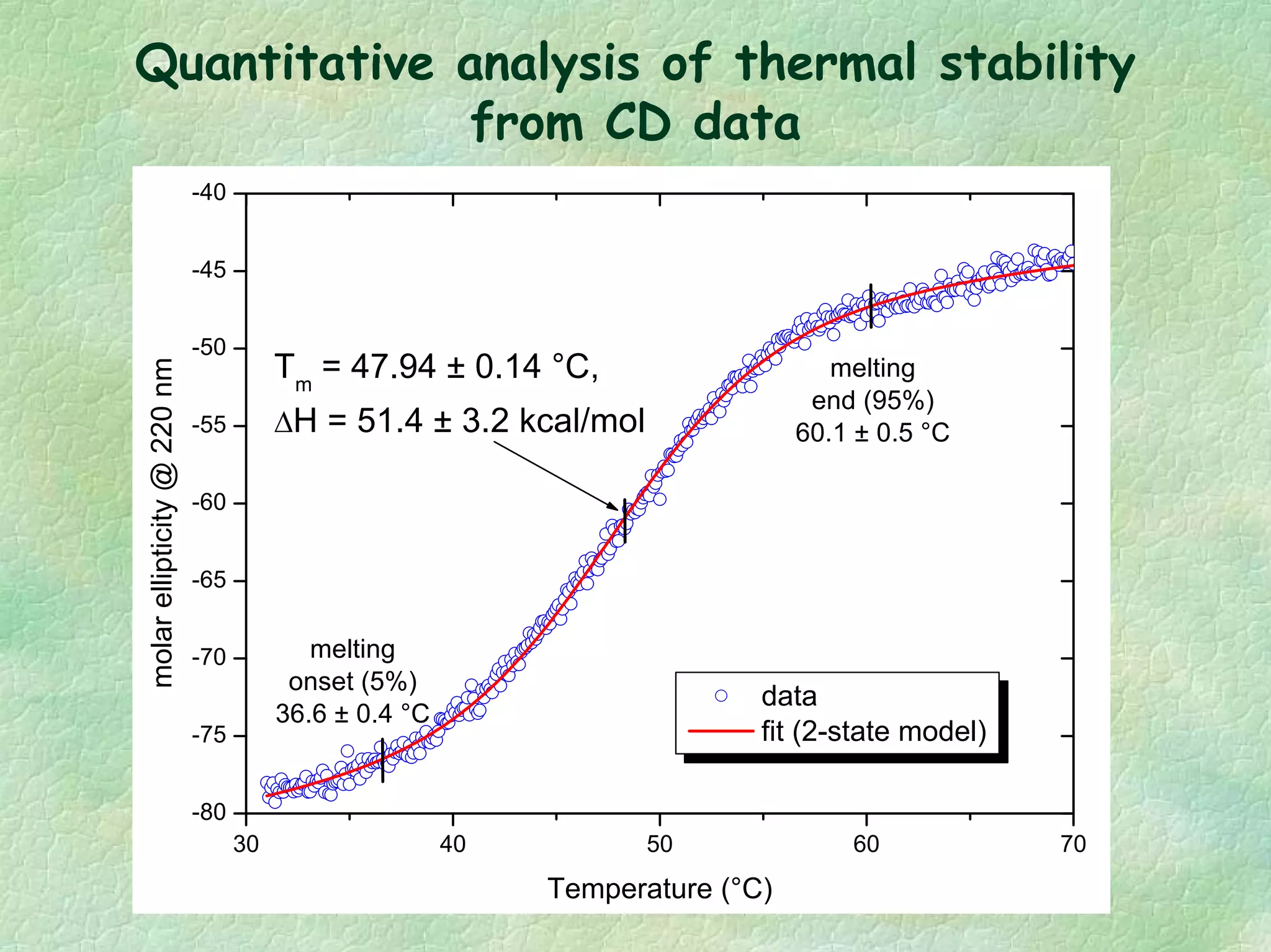
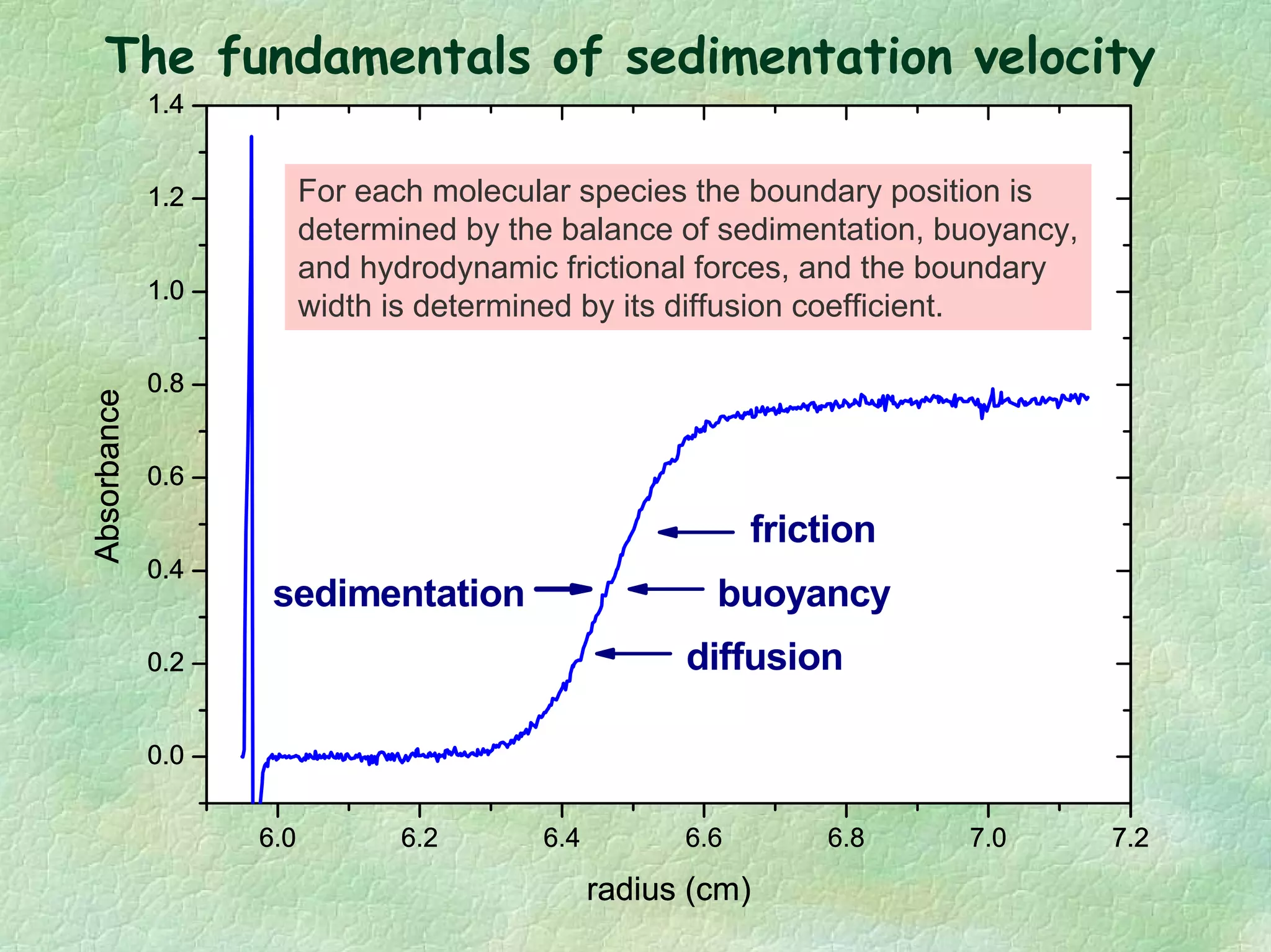
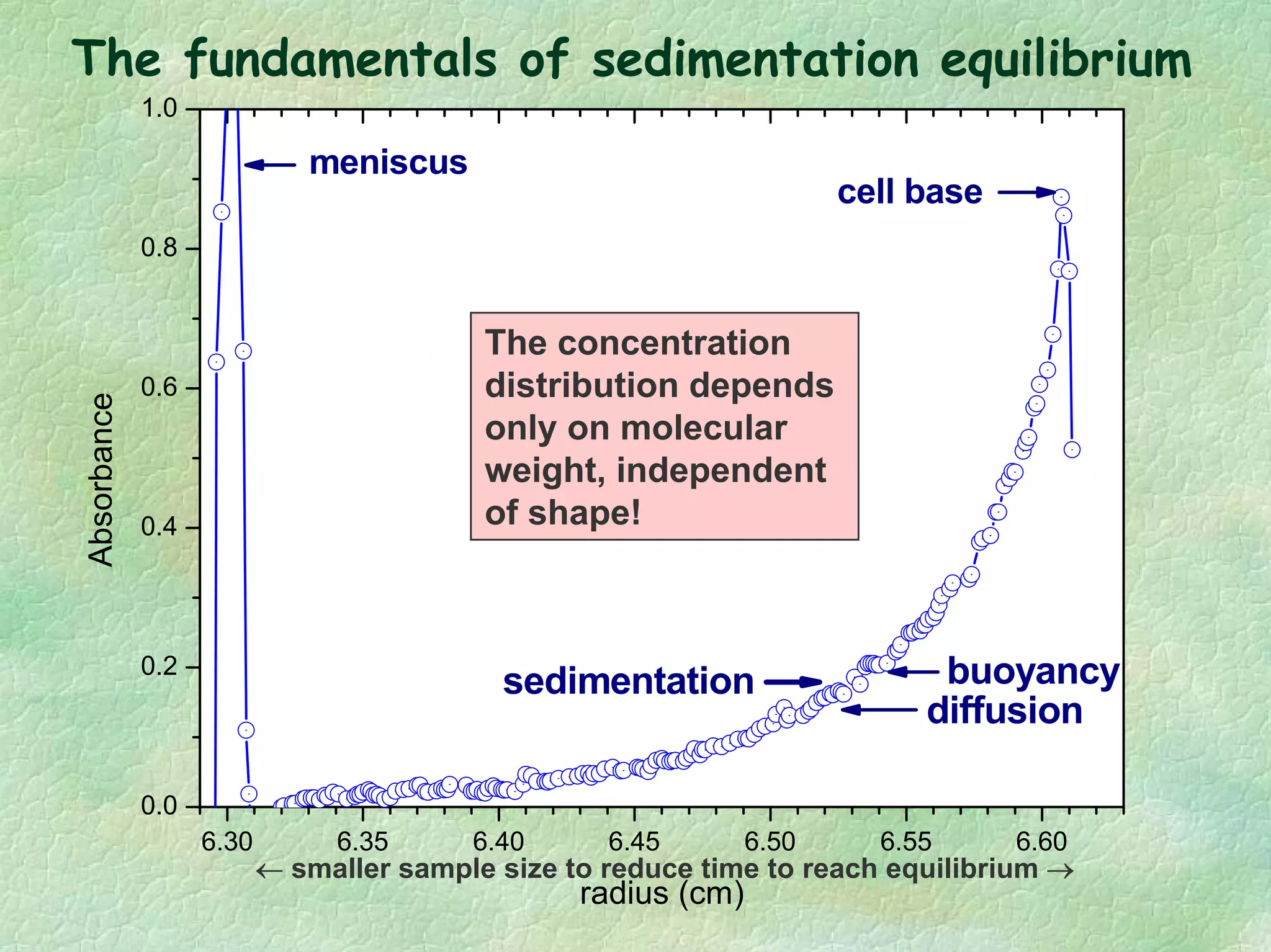
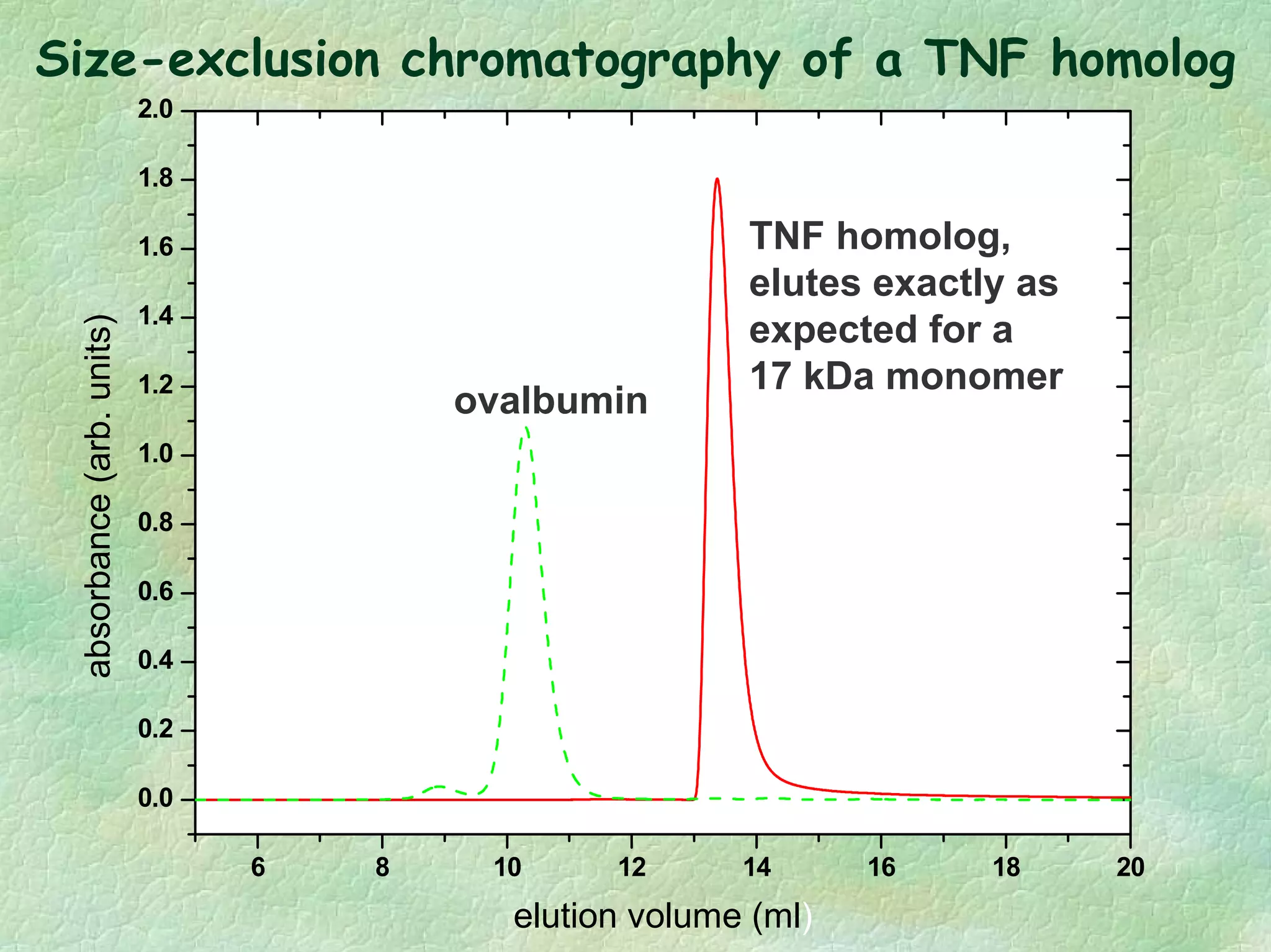
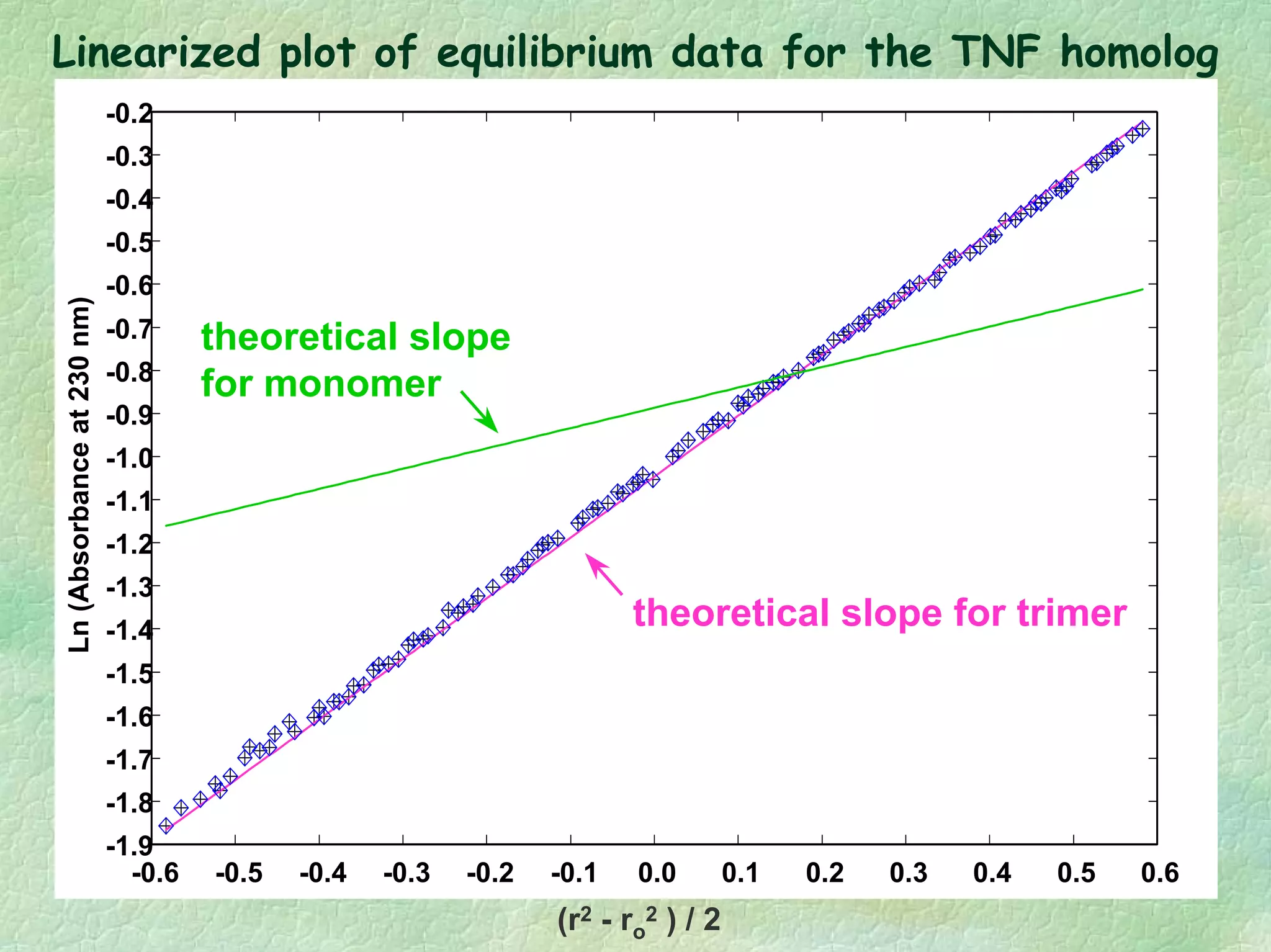
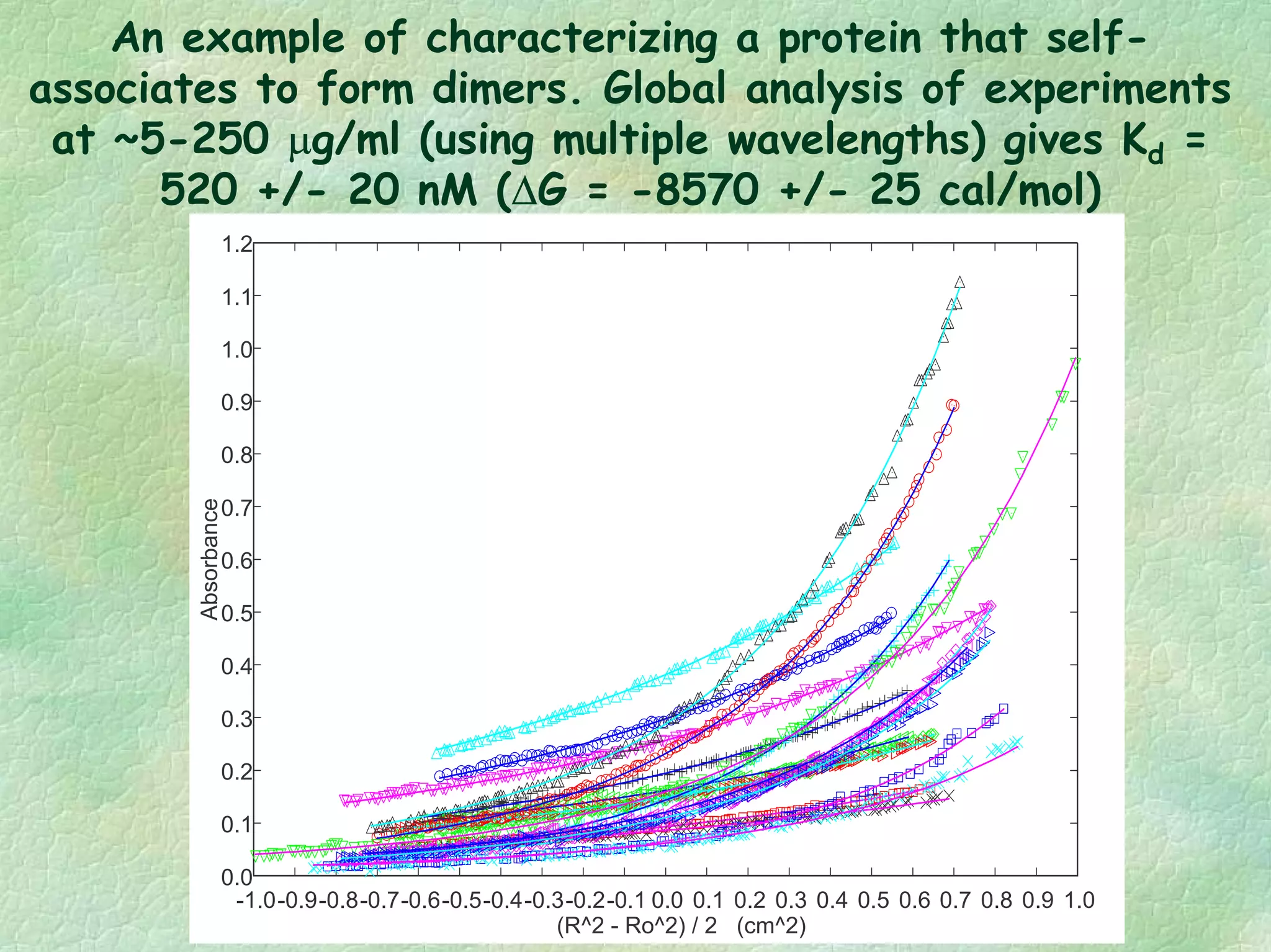
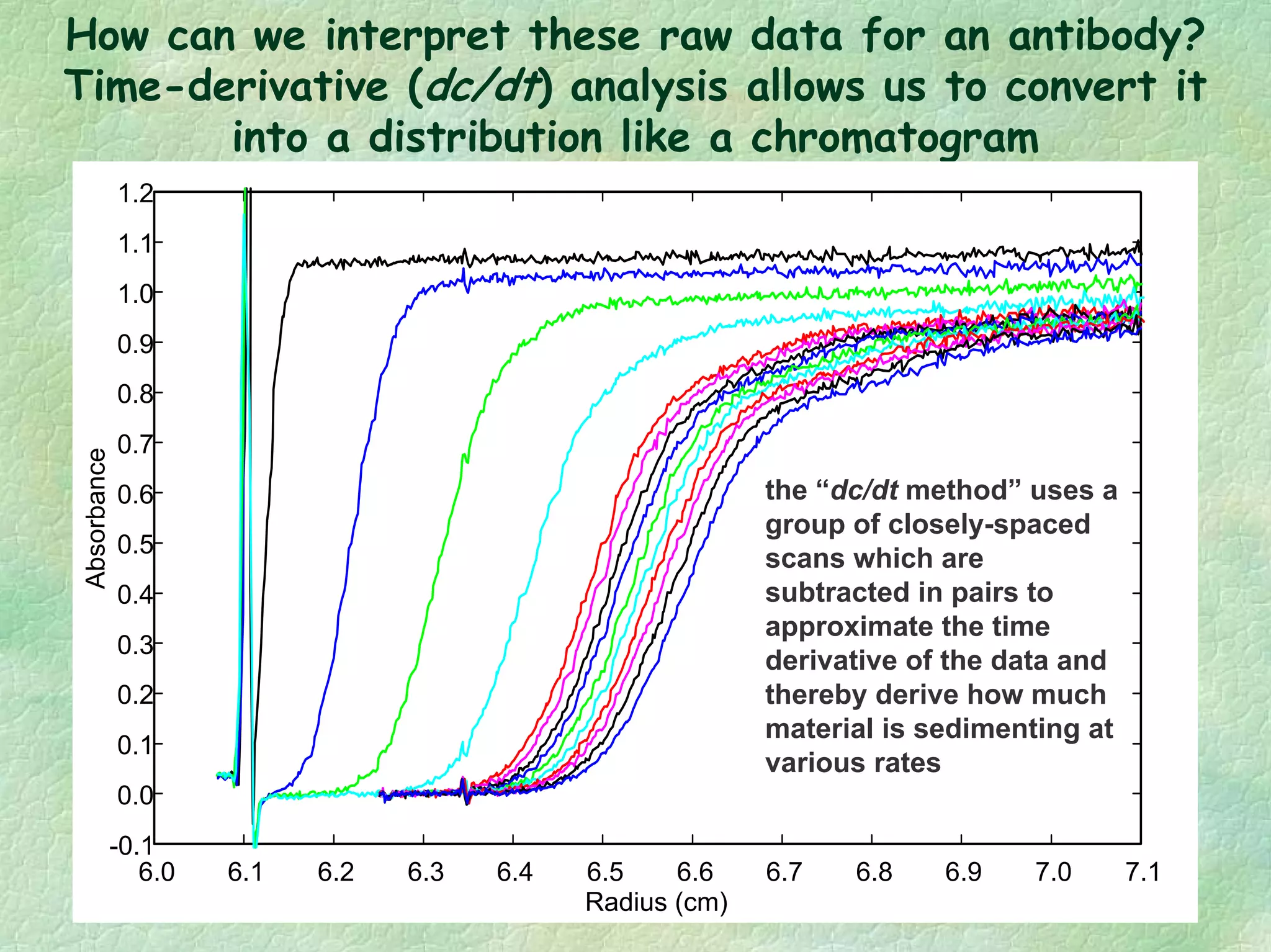
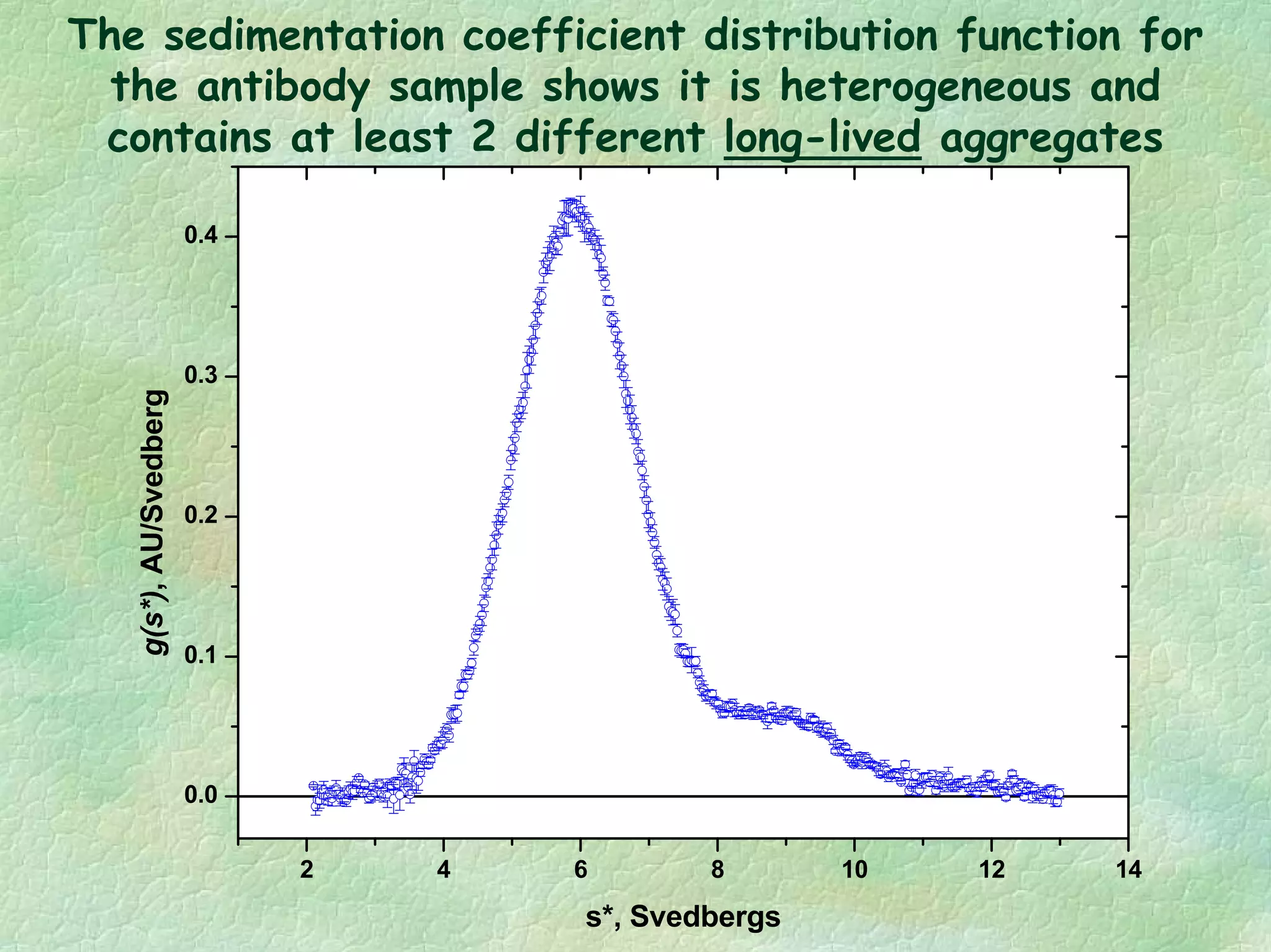
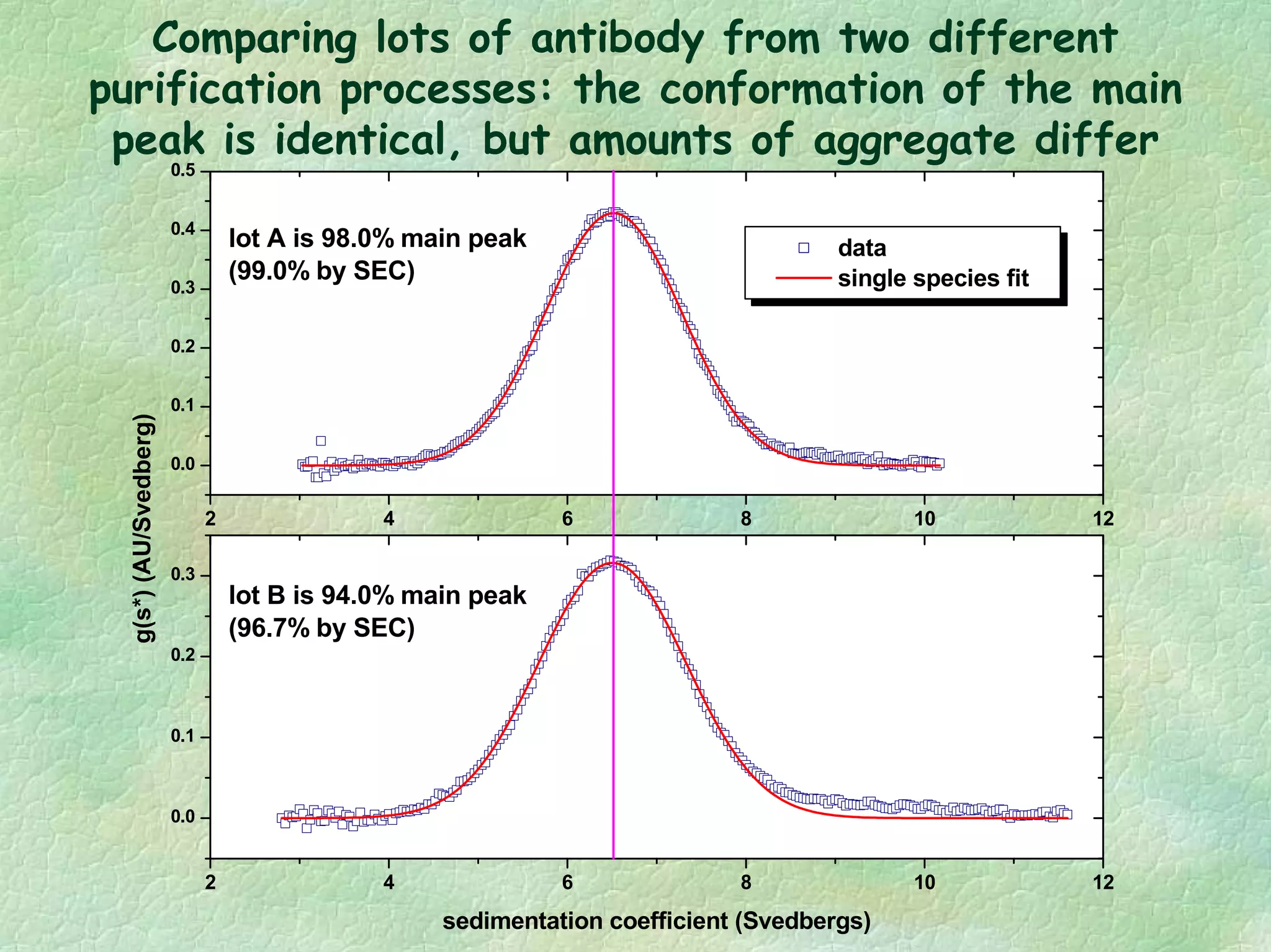
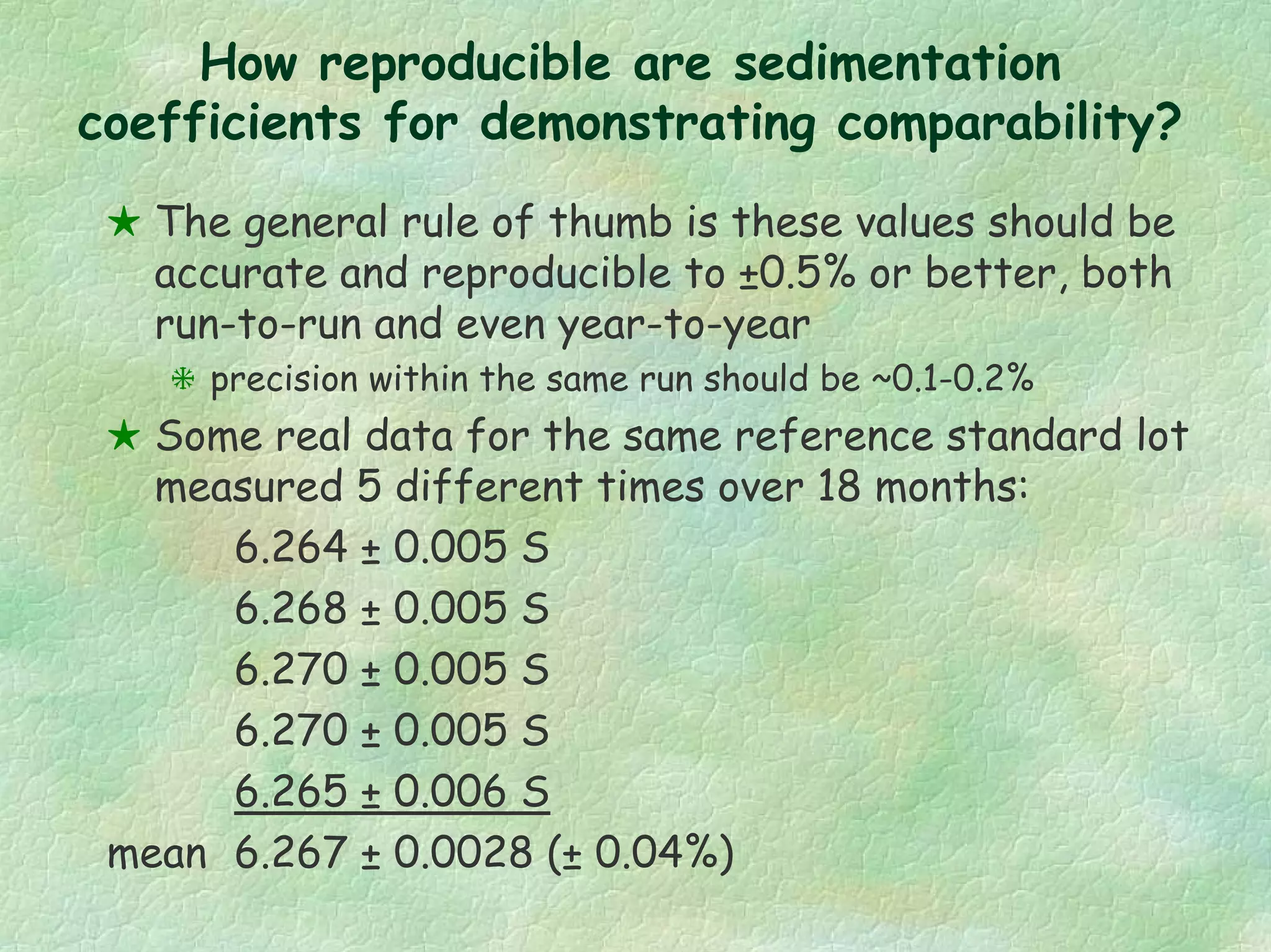
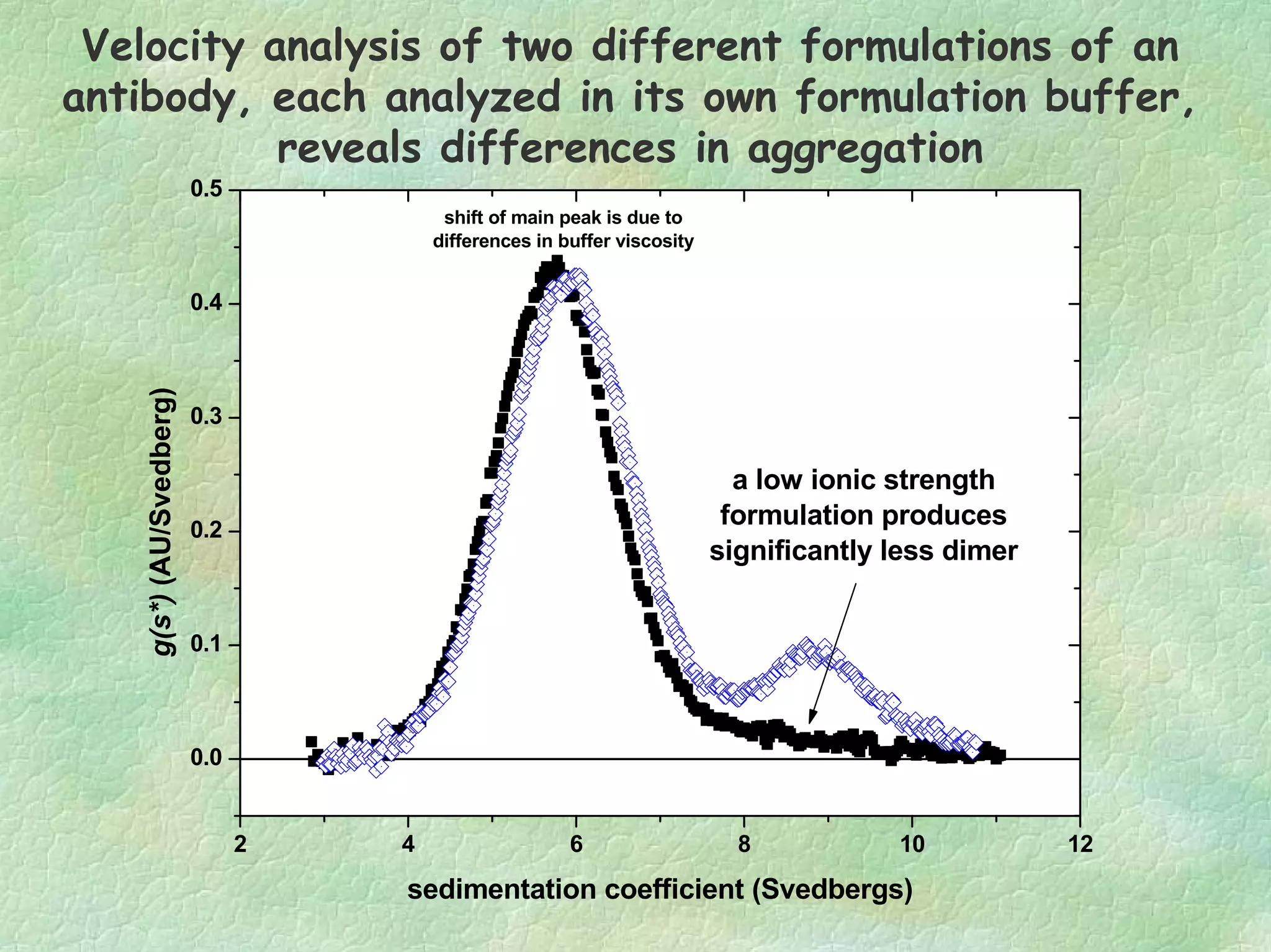
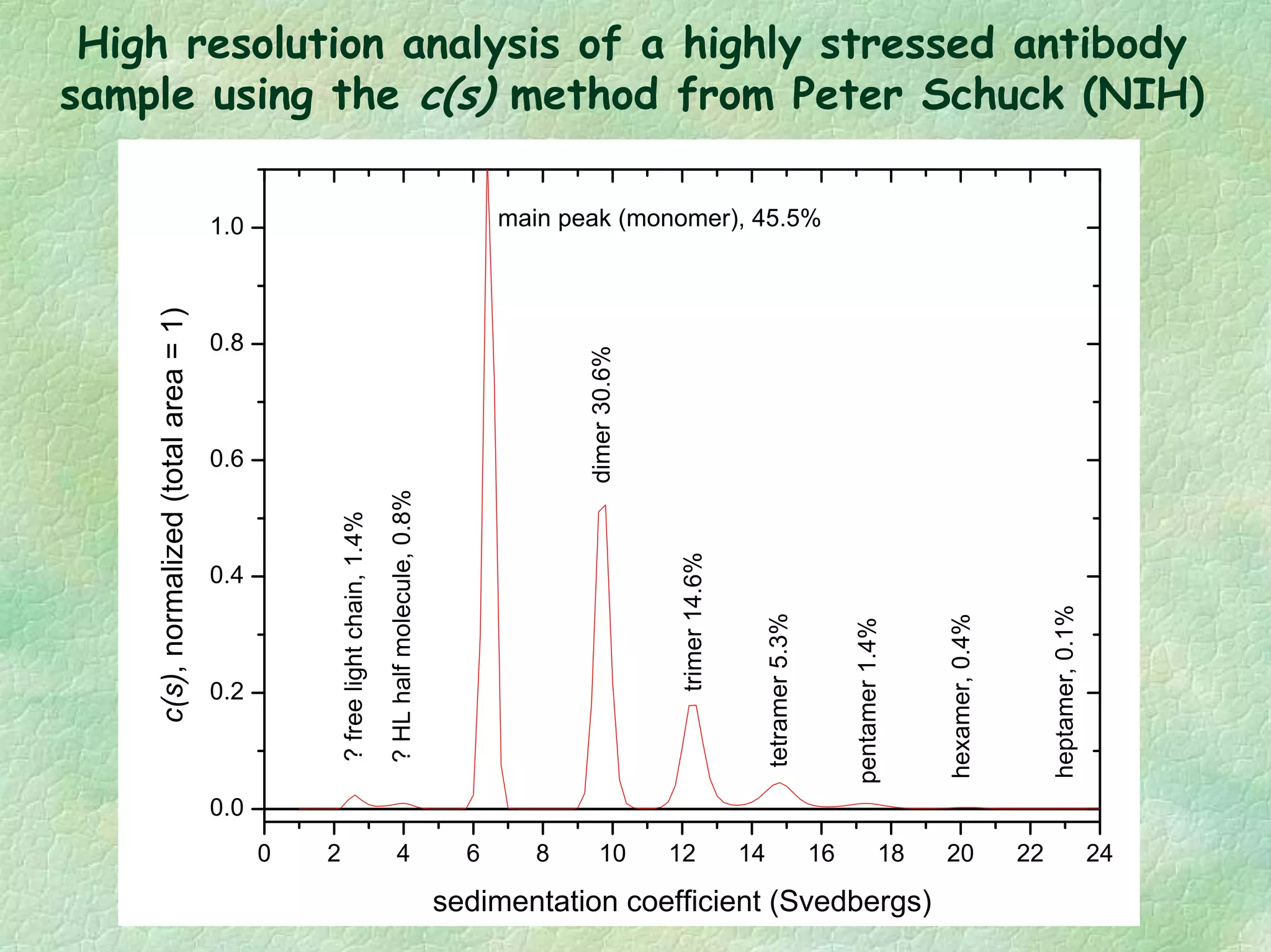
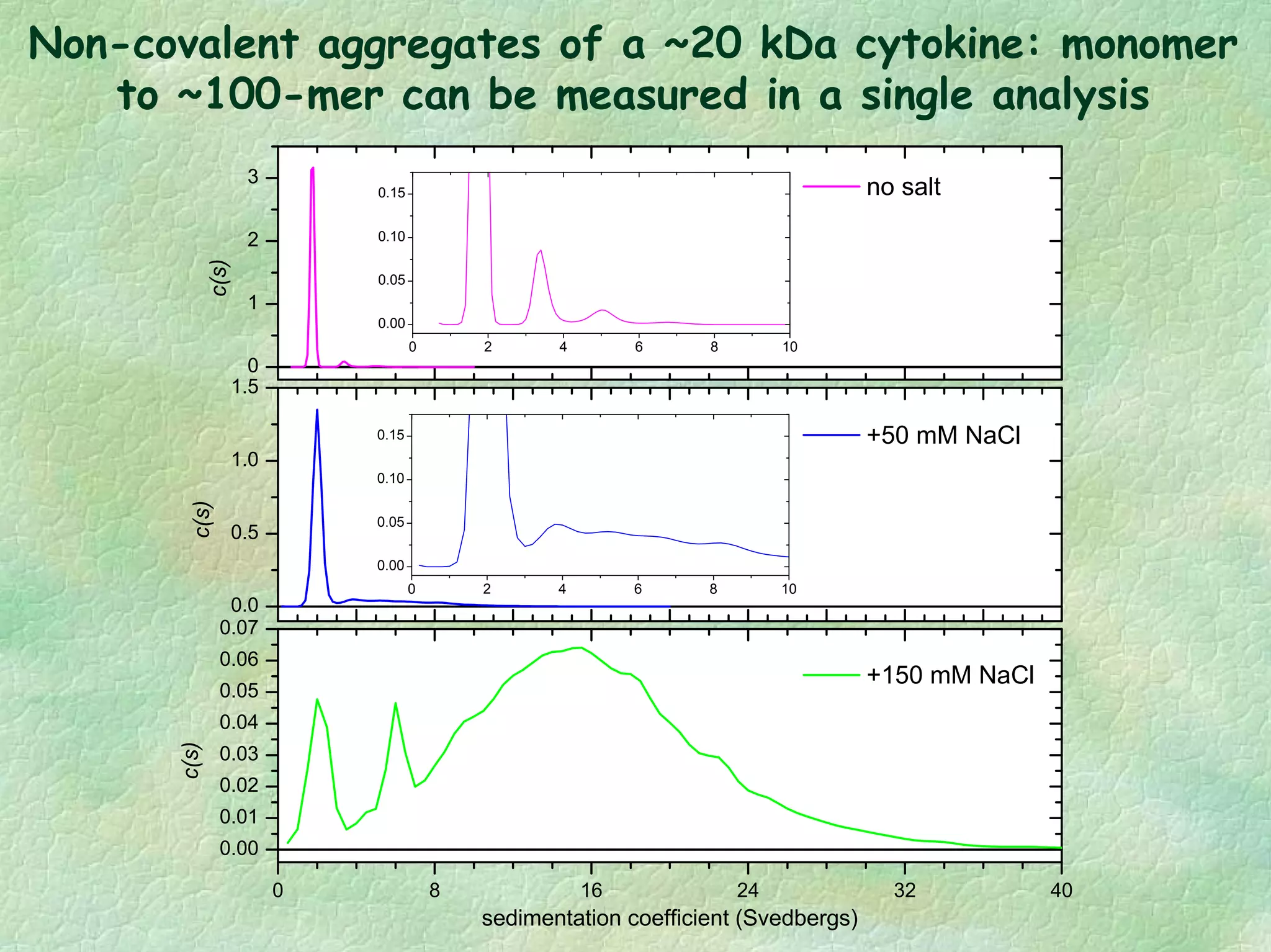
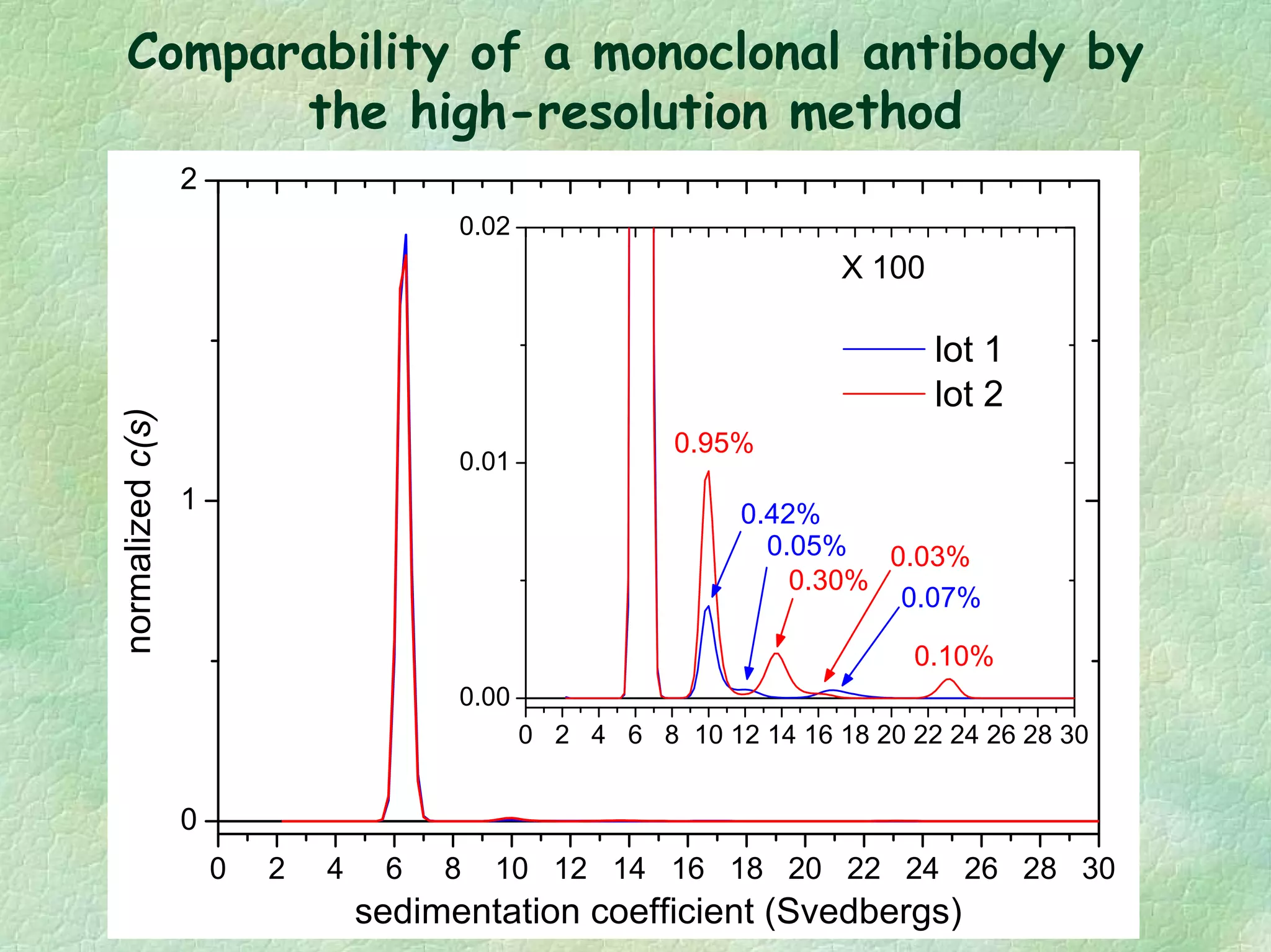
The document discusses methods for measuring protein conformational comparability using circular dichroism (CD) and analytical ultracentrifugation (AUC). It highlights the utility of CD in providing spectral fingerprints and thermal stability data, while AUC techniques offer quantitative insights into sedimentation coefficients and protein aggregation. Overall, it emphasizes the importance of these methods in characterizing protein structure and stability, essential for biopharmaceutical development.