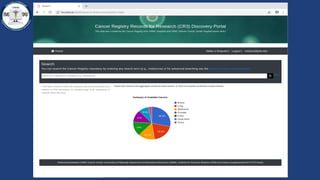
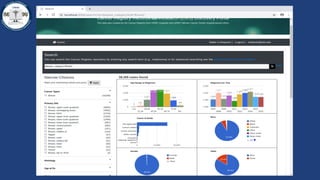
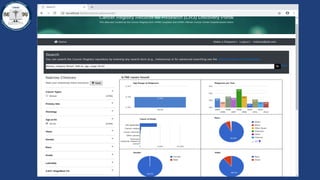
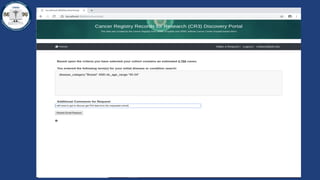
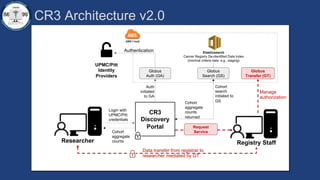
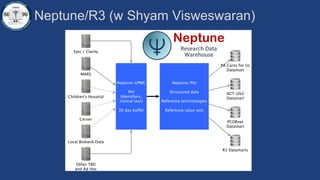
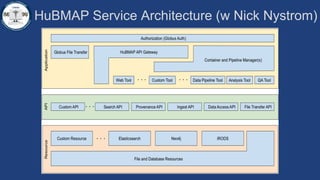
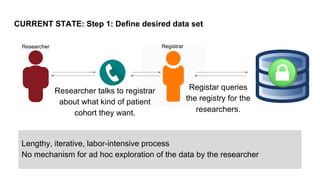
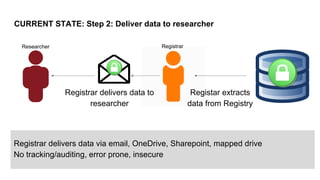
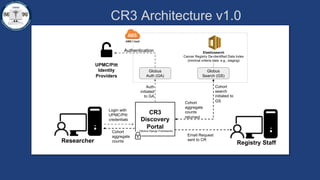
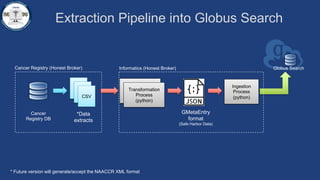
This document summarizes an effort to create an end-to-end infrastructure for cancer researchers to request, explore, and receive de-identified cancer registry data. It involves building a research portal using Globus technologies to allow researchers to search a federated index of de-identified registry data across multiple institutions while maintaining local data control. The goal is to create a scalable network of interoperable cancer registries through federation to support collaborative multi-institutional research queries.








![{
"@version": "2016-11-09",
"ingest_data": {
"@version": "2017-09-01",
"gmeta": [
{
"@version": "2017-09-01",
"subject": "https://dbmi.pitt.edu/subject/201201295:00:BRCA",
"visible_to": [
"urn:globus:groups:id:ddb13d88-d6df-11e8-9daa-0e368f3075e8"
],
"content": {
"clinical_t": "T2",
"mets_at_dx": "No distant metastasis",
"recurrence": "No",
"vitals": "Alive",
"clinical_n": "N0",
"race": "White",
"histology": "Infiltrating duct carcinoma, NOS",
"path_stage": "2A",
"facility": "Magee",
"histology_code": "85003",
"disease_category": "Breast",
"dx_age_range": "35-44",
"grade": "Grade II: Mod diff, mod well diff,",
"spanish_hispanic_origin": "Non-Spanish; non-Hispanic",
"year_last_contact": "2017",
"cs_stage": "2A",
"site": "Breast, NOS",
"cs_extension": "100",
"clinical_stage": "2A",
"cause_of_death": "Not applicable",
"path_n": "N0",
"path_t": "T2",
"cancer_status": "No evidence of this tumor",
"age_at_dx": "36",
"gender": "Female",
"summary_stage": "Localized",
"clinical_m": "M0",
"laterality": "Left - origin of primary",
"dx_year": "2012",
"site_code": "C509"
}
}
]
}
}
GMetaEntry data in GS
visible_to:
content:
(scope limited to defined
Globus Group)](https://image.slidesharecdn.com/10silversteinupittinformatics-190515221207/85/Health-Sciences-Research-Informatics-Powered-by-Globus-17-320.jpg)