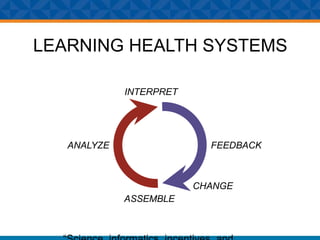
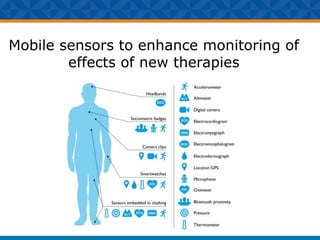
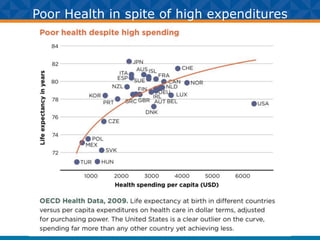
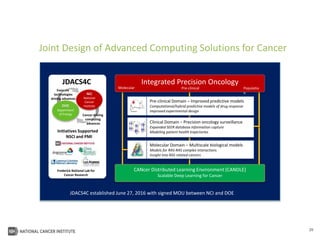
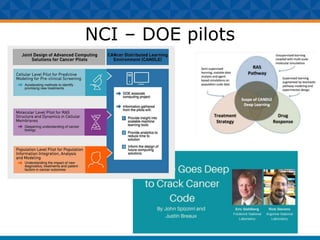
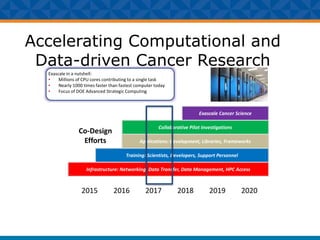
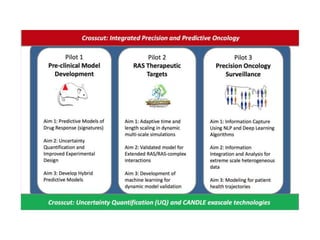
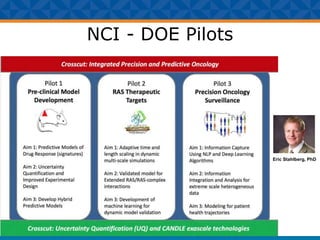
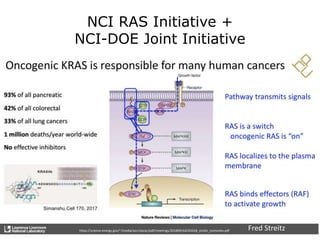
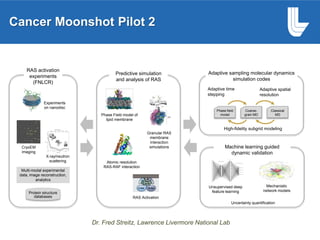
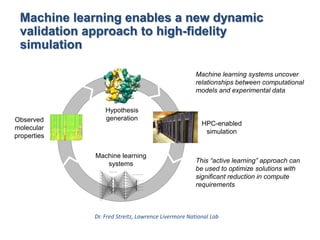
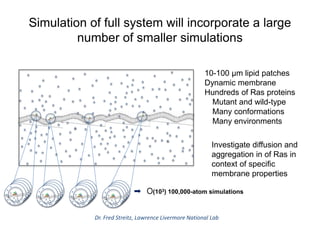
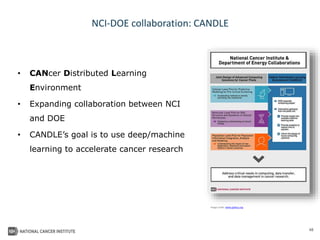
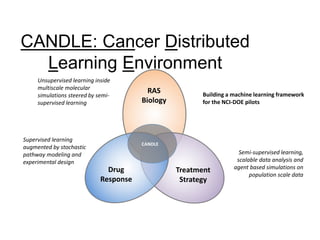
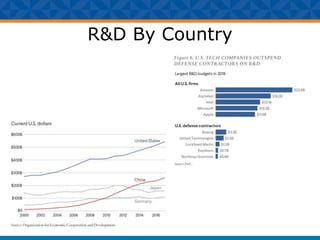
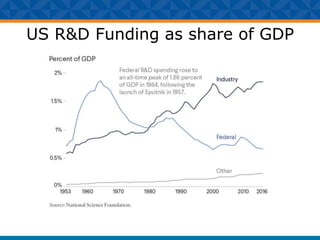
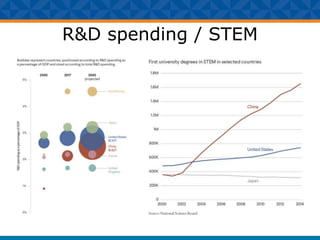
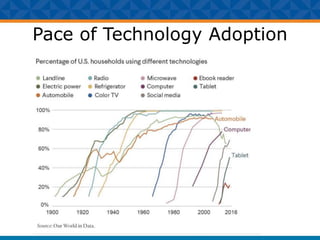
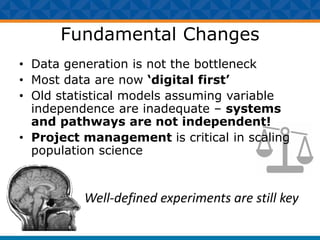
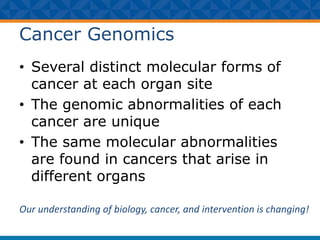
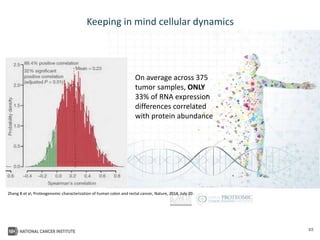
The document outlines Dr. Warren A. Kibbe's insights on the role of computing and data in precision oncology, emphasizing the need for advanced computational models and collaboration between the National Cancer Institute and the Department of Energy. It discusses the challenges of cancer research, the importance of data science, and ongoing initiatives like the Cancer Moonshot and Precision Medicine Initiative aimed at improving cancer treatment and understanding through big data and machine learning. Kibbe highlights the urgency of addressing cancer as a grand challenge in health care, with a focus on leveraging technology to enhance patient outcomes and research capabilities.