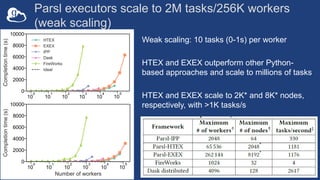
The document discusses challenges in research data management and analysis, highlighting issues such as inefficiencies, errors, and collaboration difficulties within large-scale science. It presents various tools and platforms developed by Globus Labs, including FuncX, Parsl, Polyner, and Ripple, aimed at automating research workflows and enhancing data accessibility and usability. The document emphasizes the importance of high-performance computing and the integration of distributed resources to optimize scientific analysis and improve research outcomes.