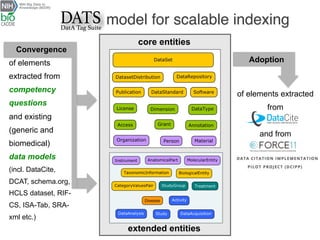
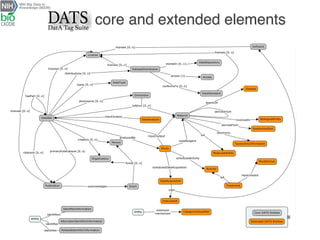
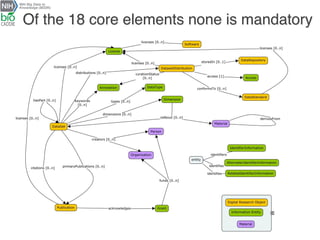
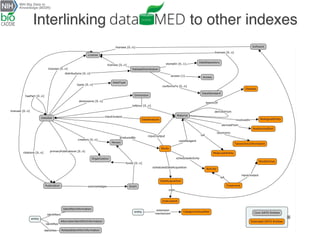
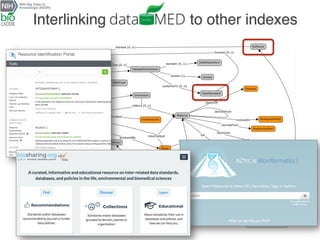
The document describes the DATS (Data Tag Suite) model, which aims to provide a standardized way to index datasets through a community effort. The DATS model was developed by combining use cases and existing data schemas, and represents datasets and their metadata in a scalable way. It focuses on key descriptors like authors, datasets, publications, and funding to enable discoverability. The DATS model is serialized in JSON and JSON-LD using schema.org to increase visibility, accessibility, and search engine ranking. It is being adopted by databases and aligned with other bioscience metadata efforts.