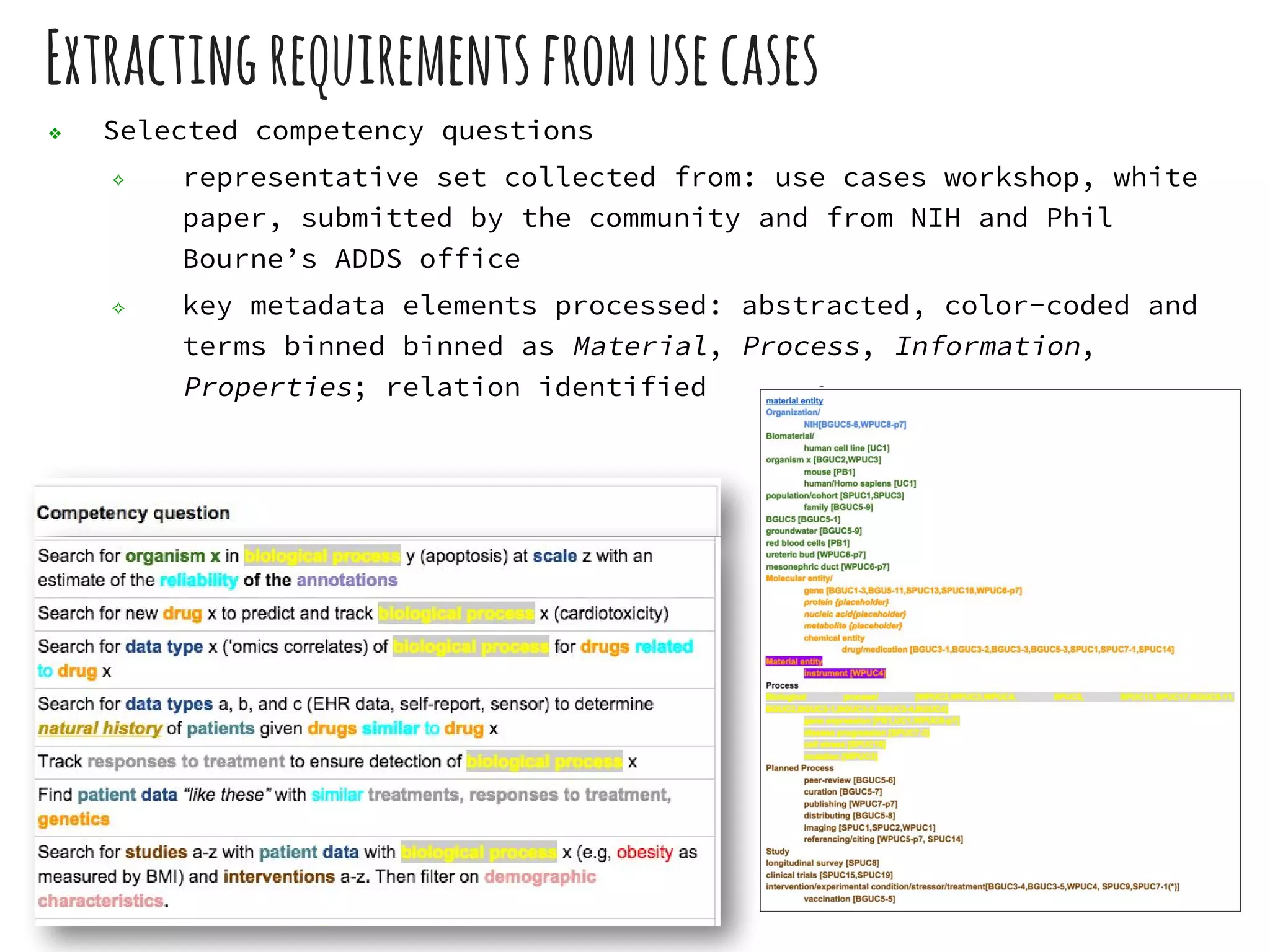
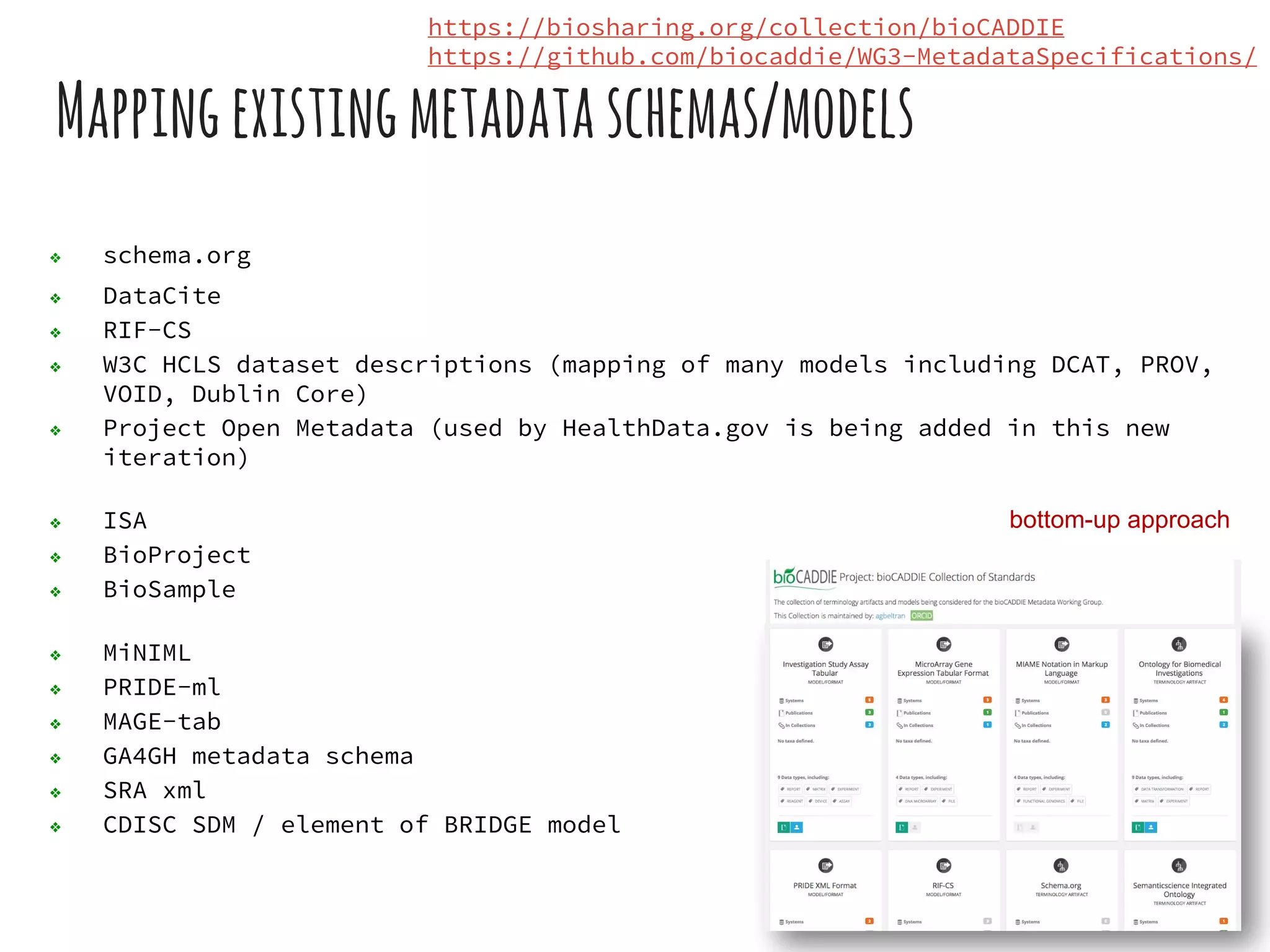
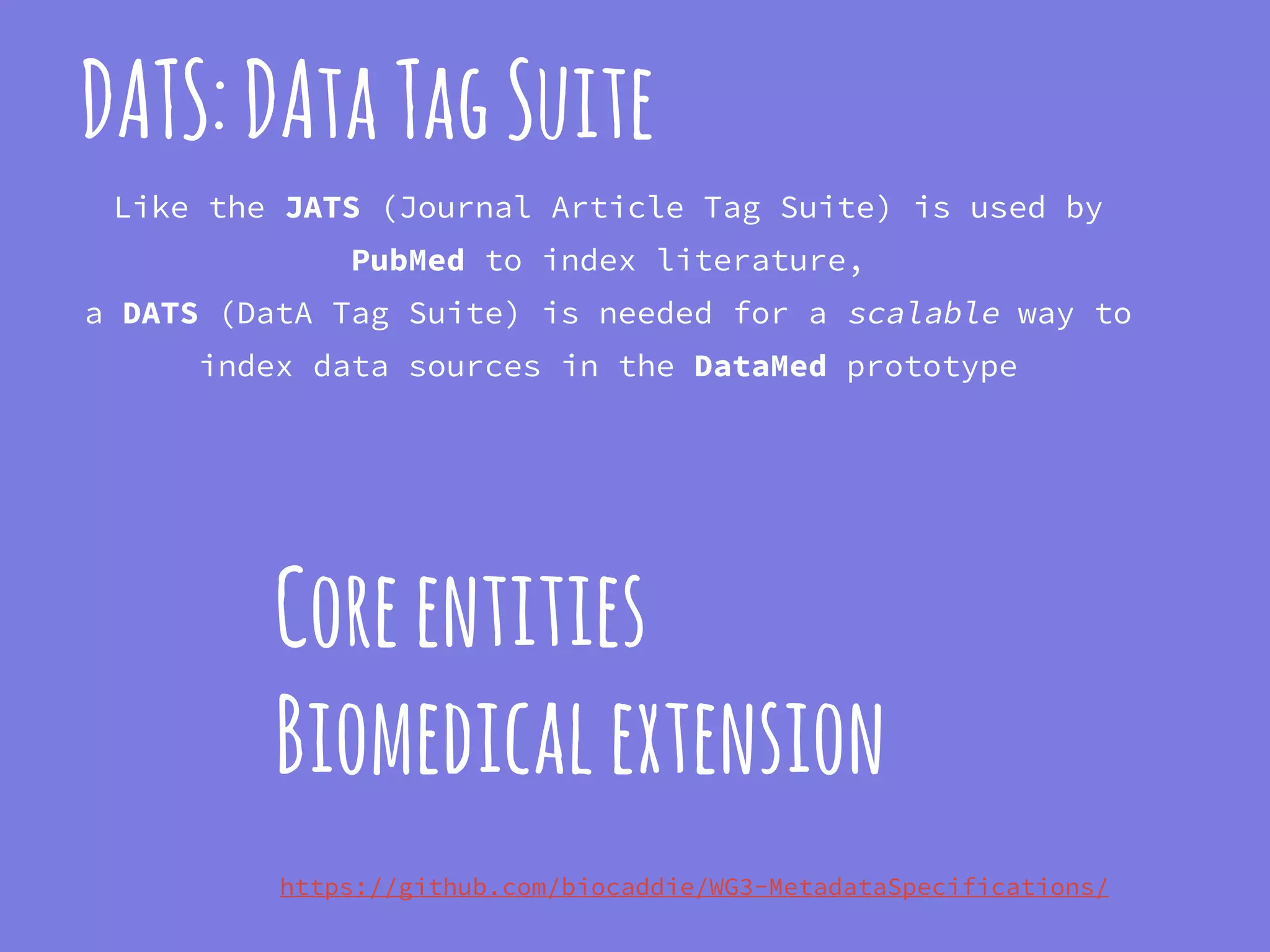
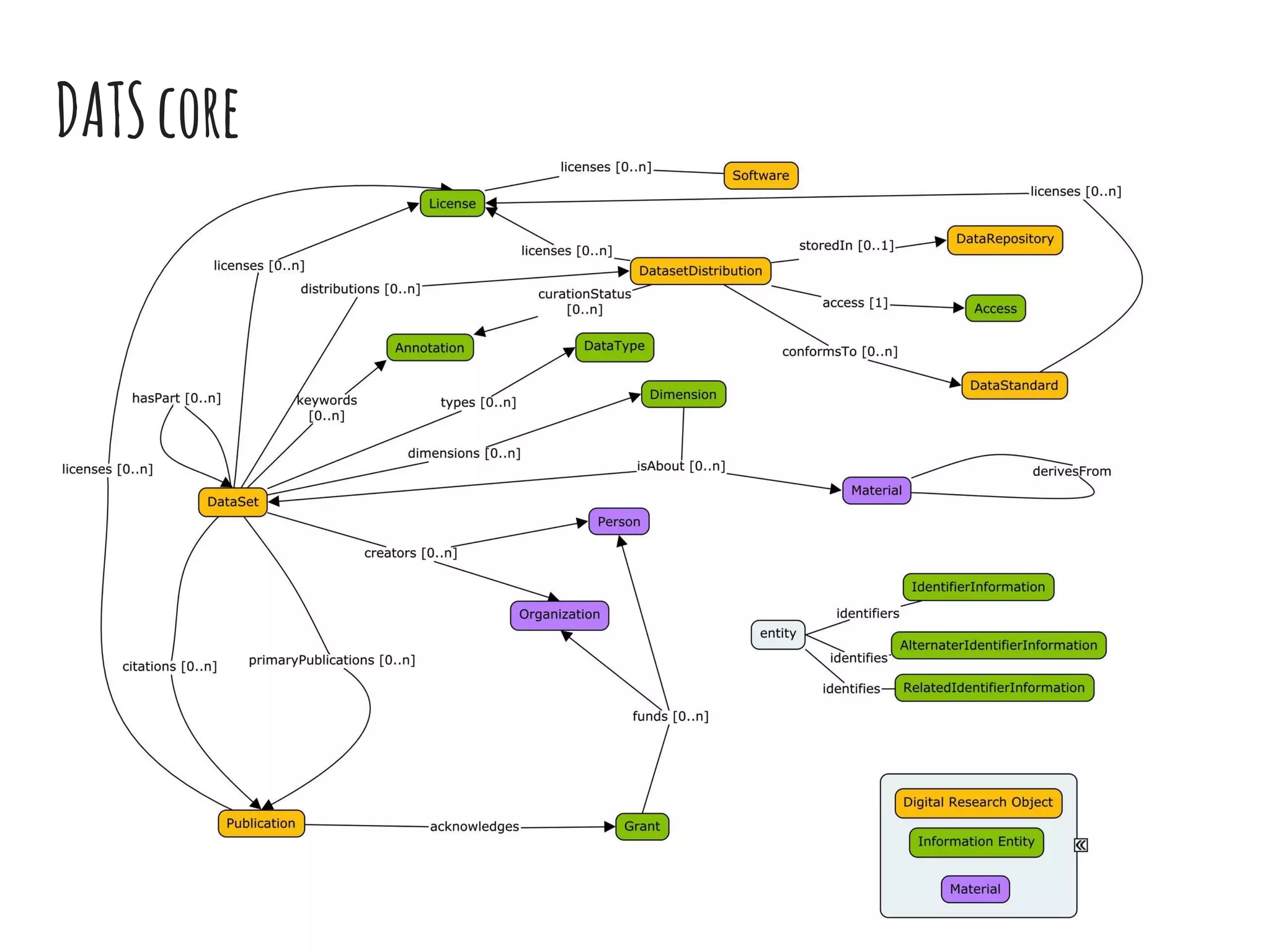
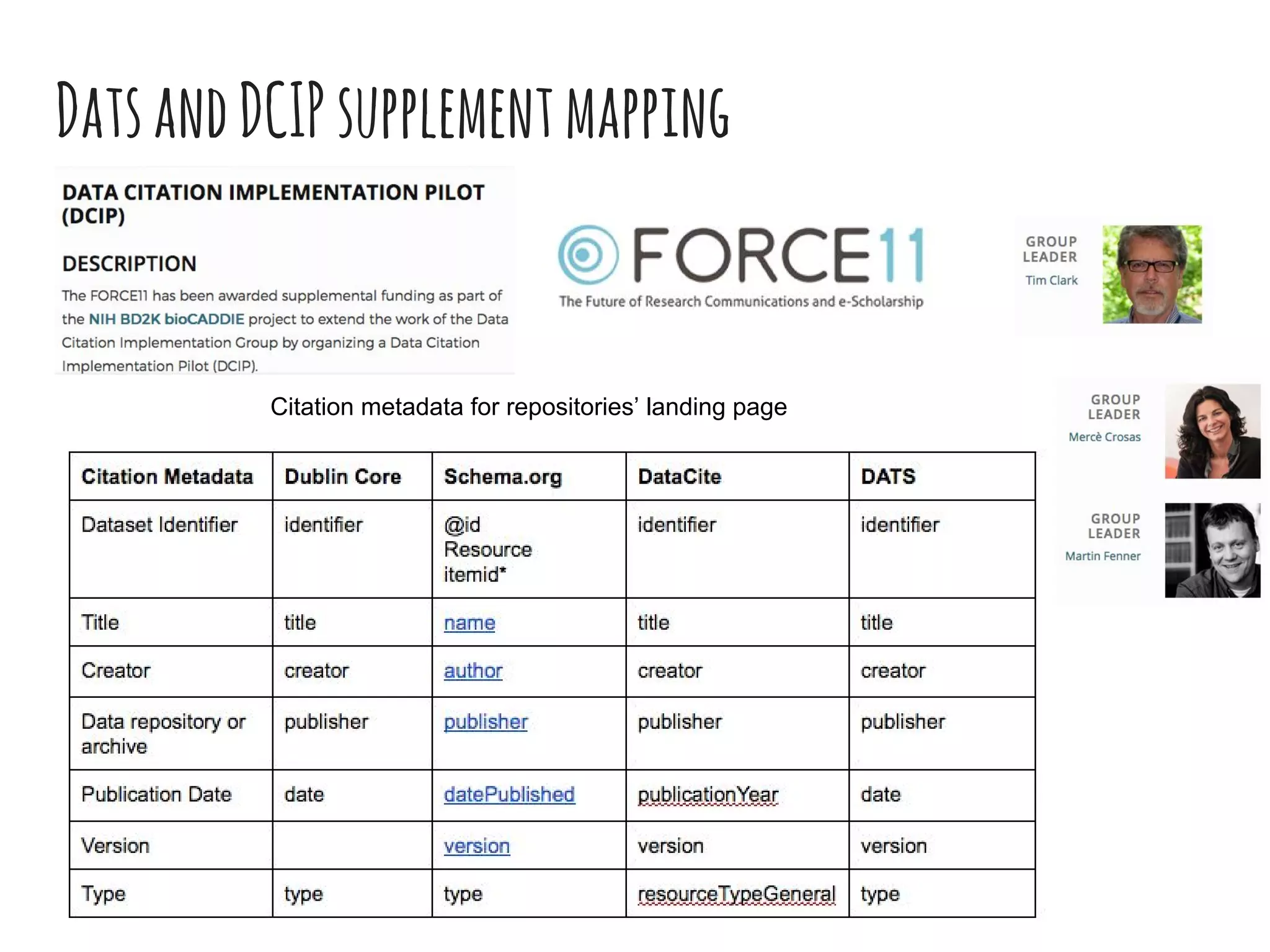
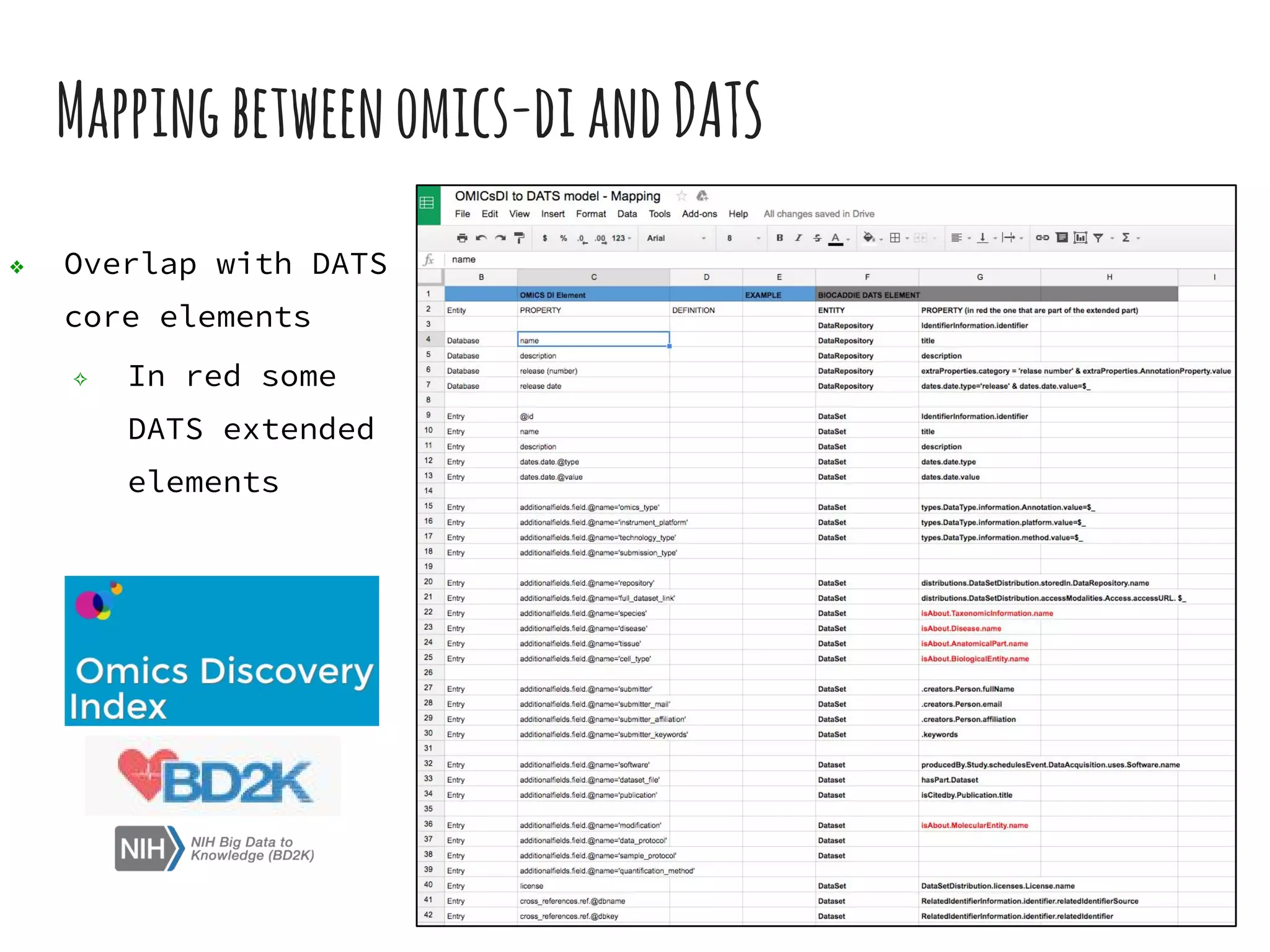
The document discusses the development of the DATS (Data Tag Suite) model for describing scientific datasets to enable data discovery. Key points:
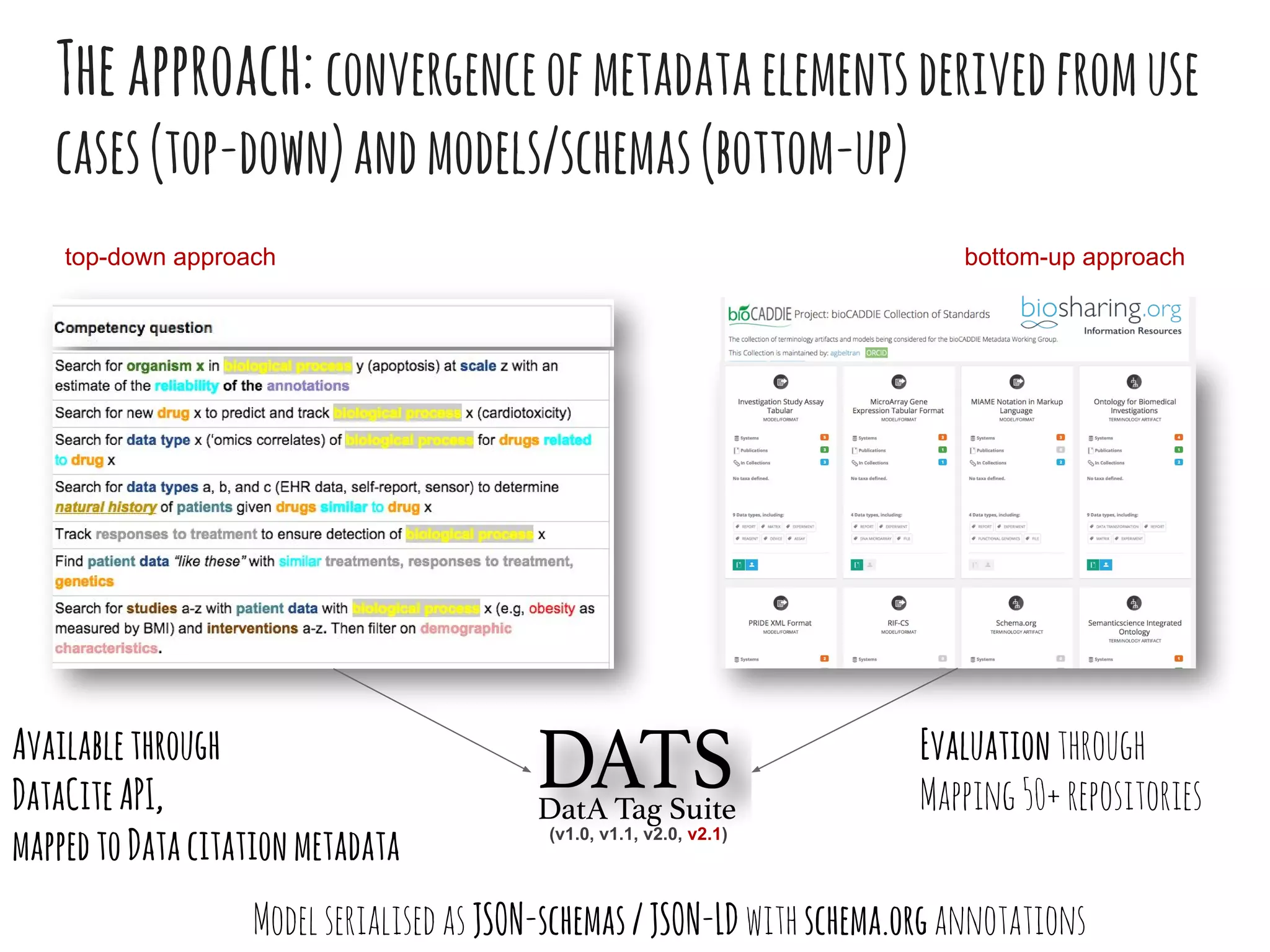
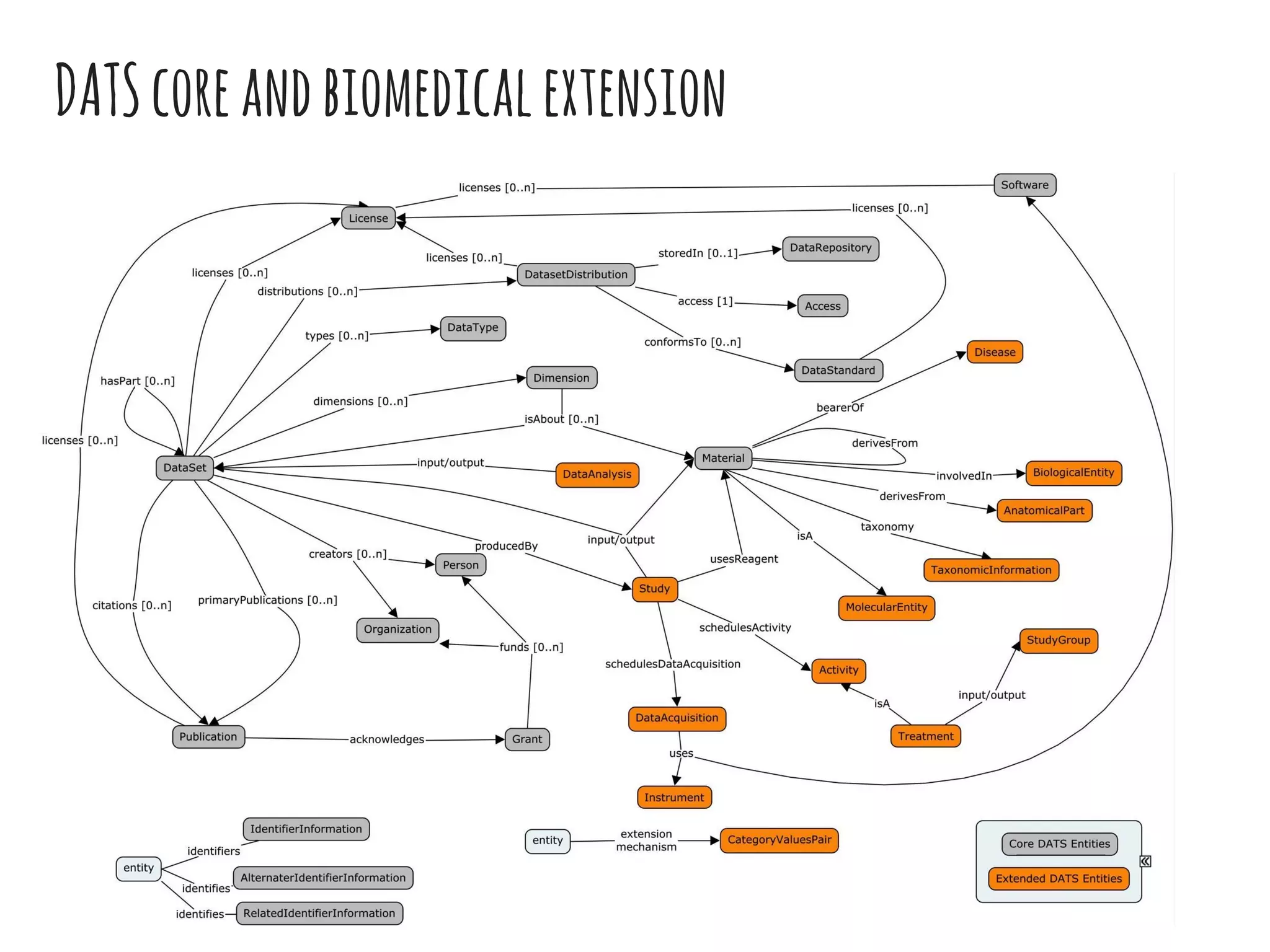
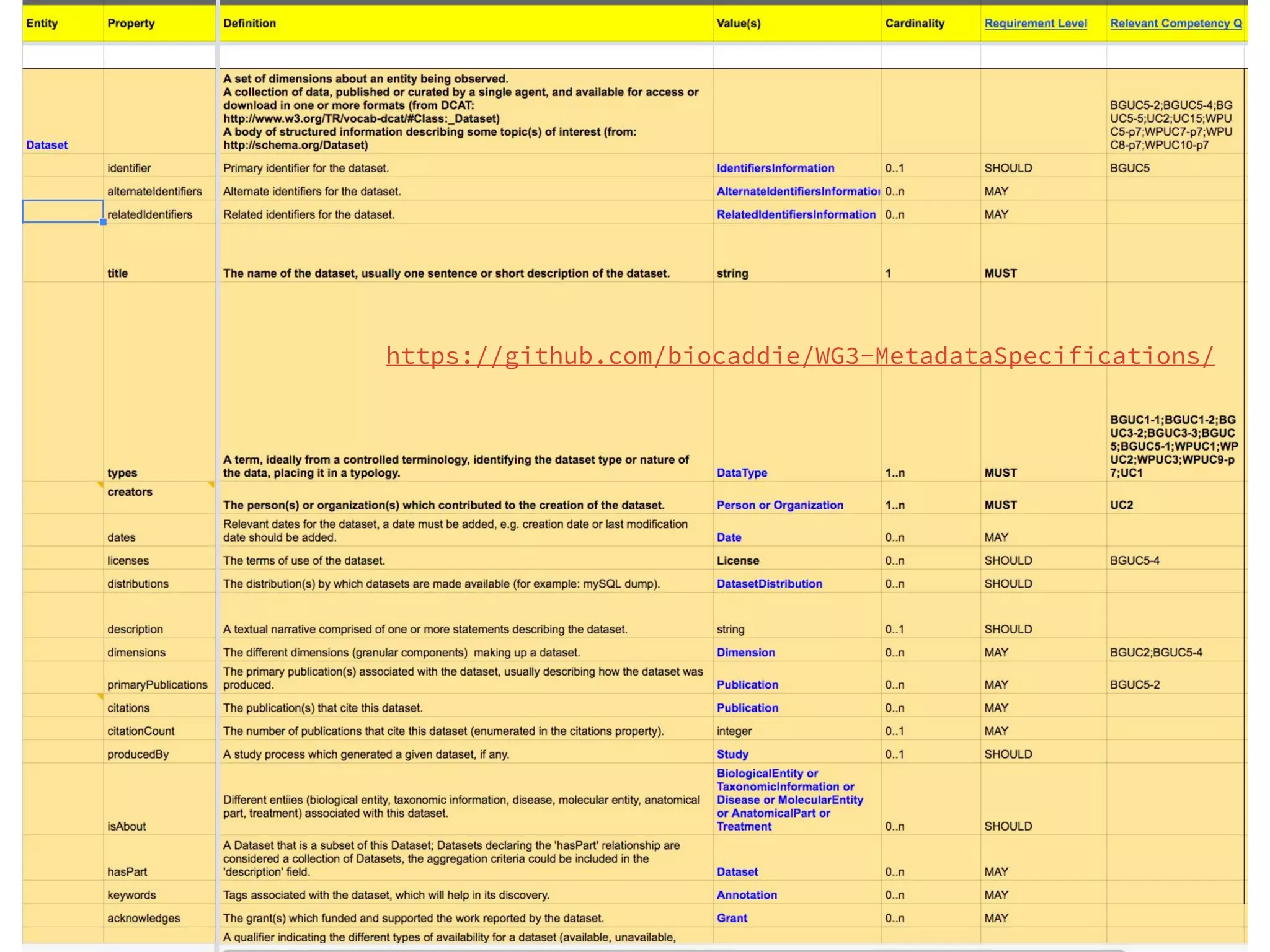
- DATS is designed around the core entity of a "dataset" and aims to converge metadata elements from use cases and existing schemas/models in a top-down and bottom-up approach.
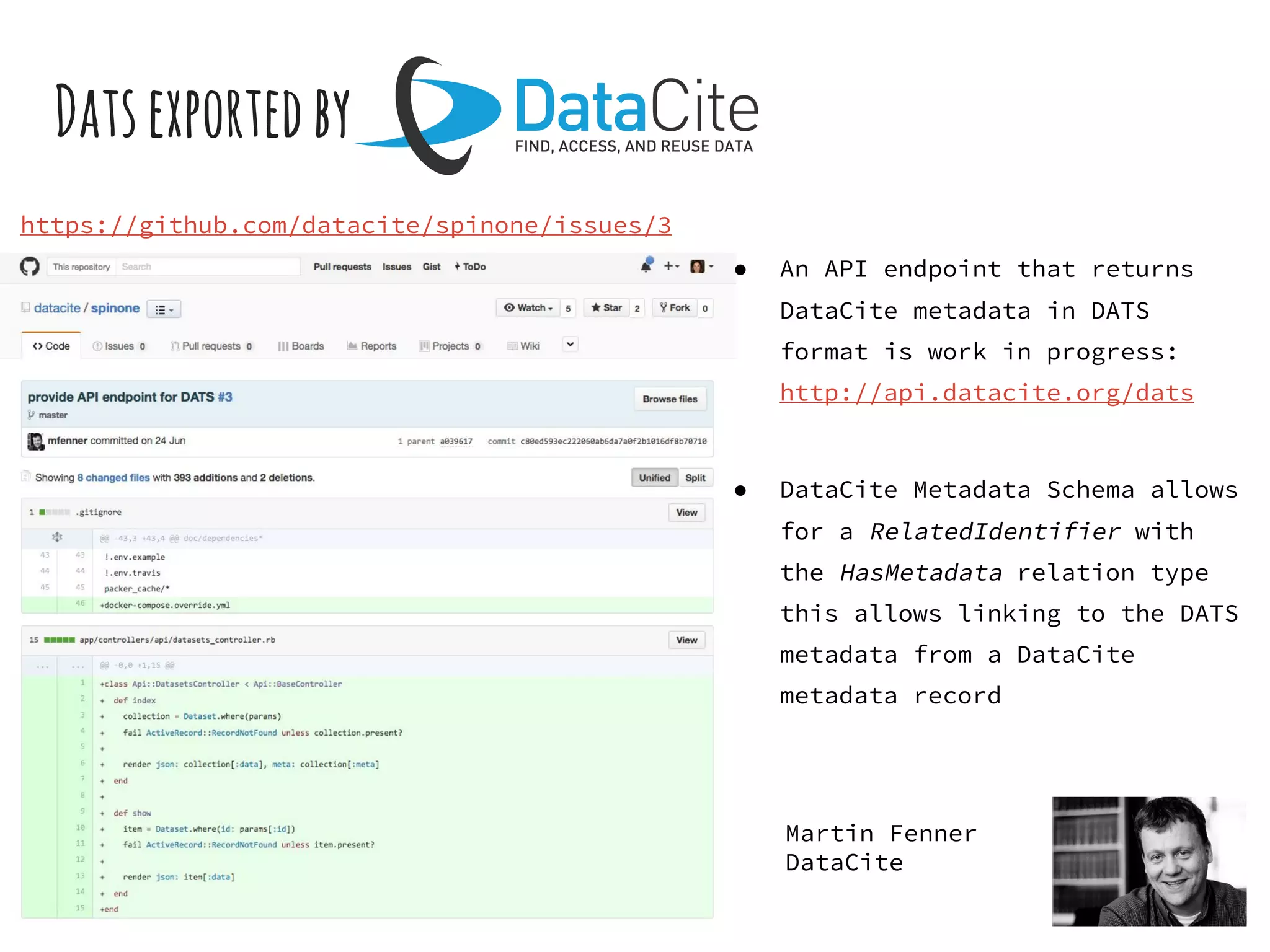
- The DATS model and a biomedical extension have been developed through a community-driven process and are available on GitHub.
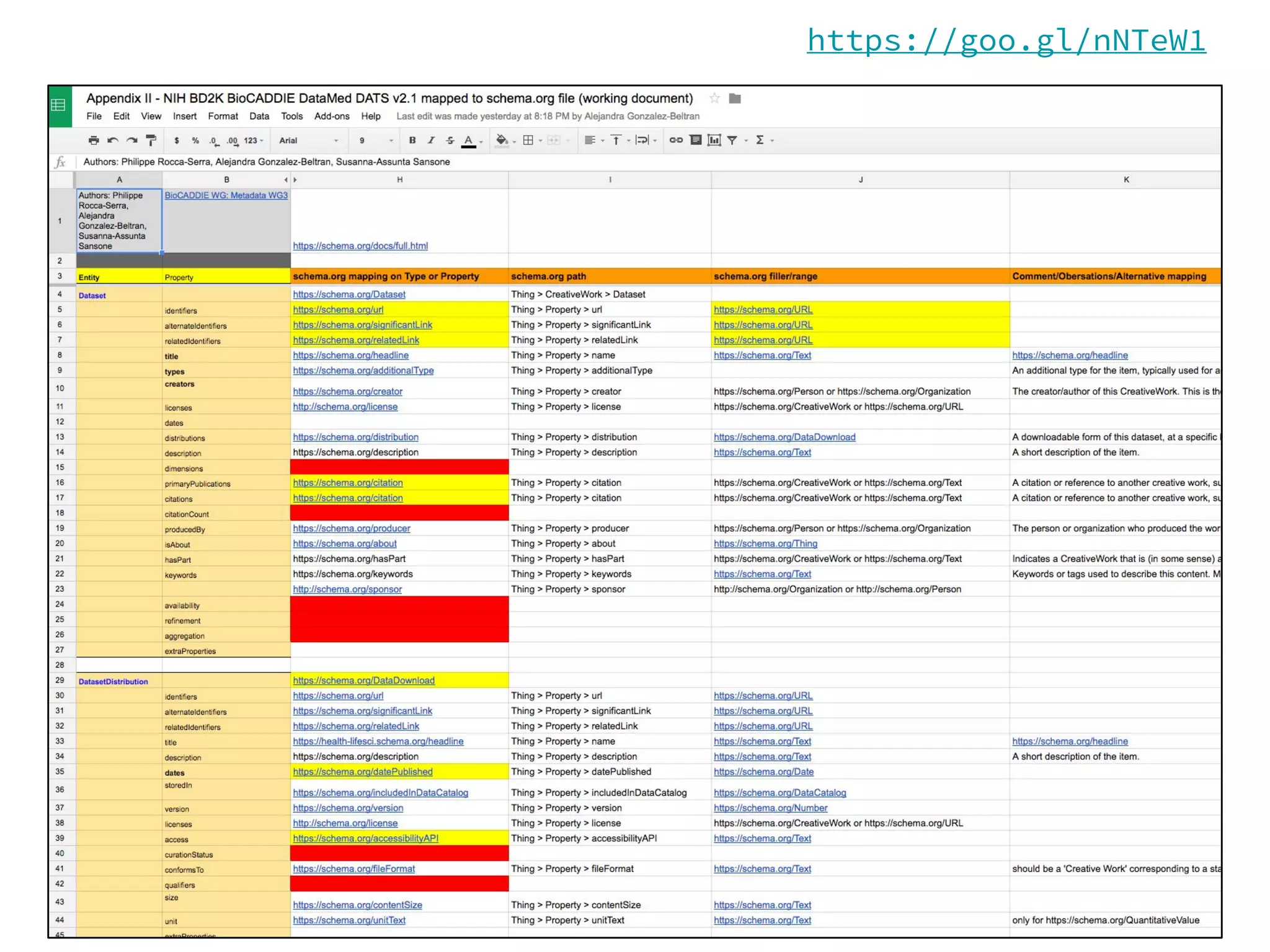
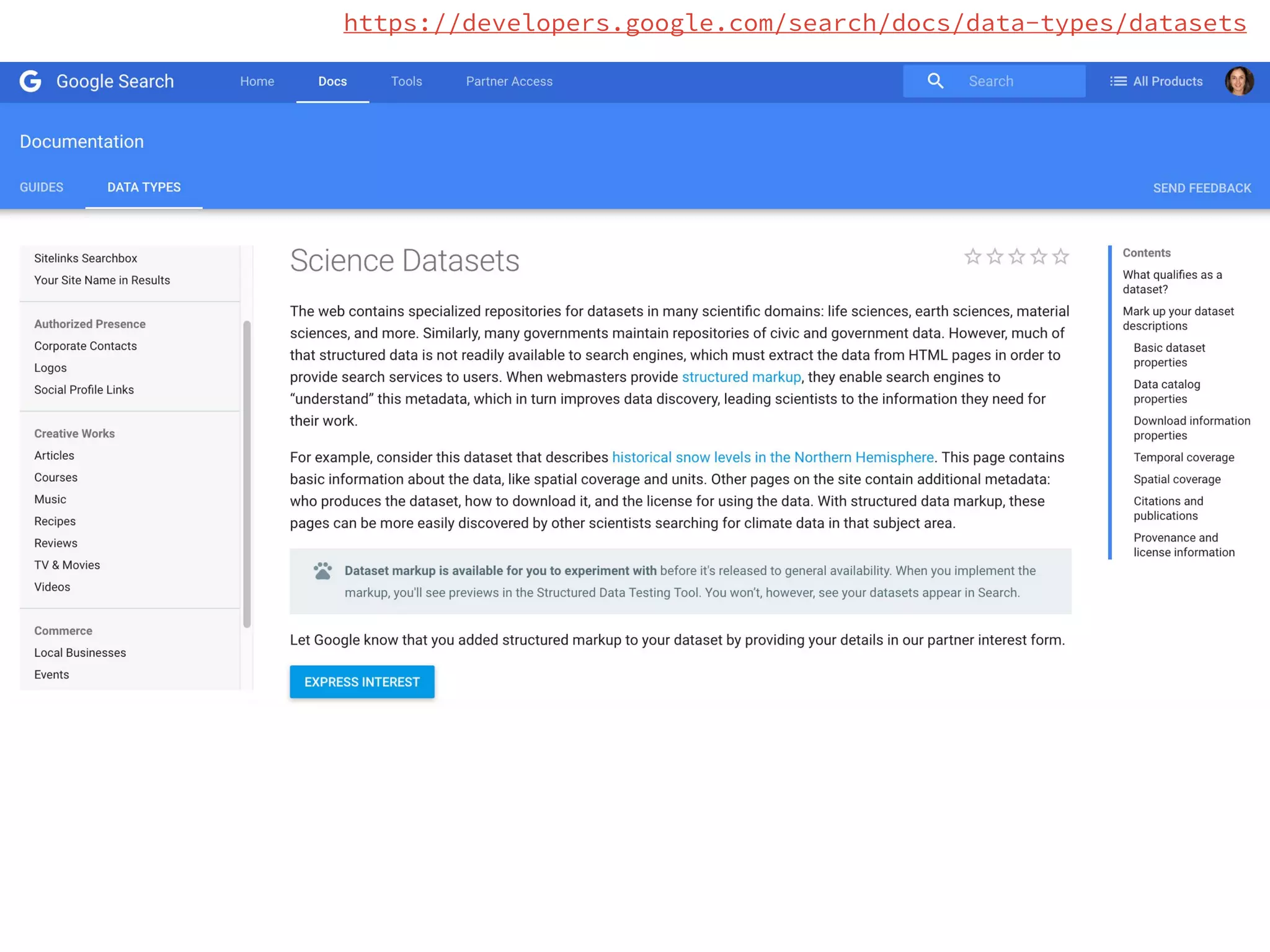
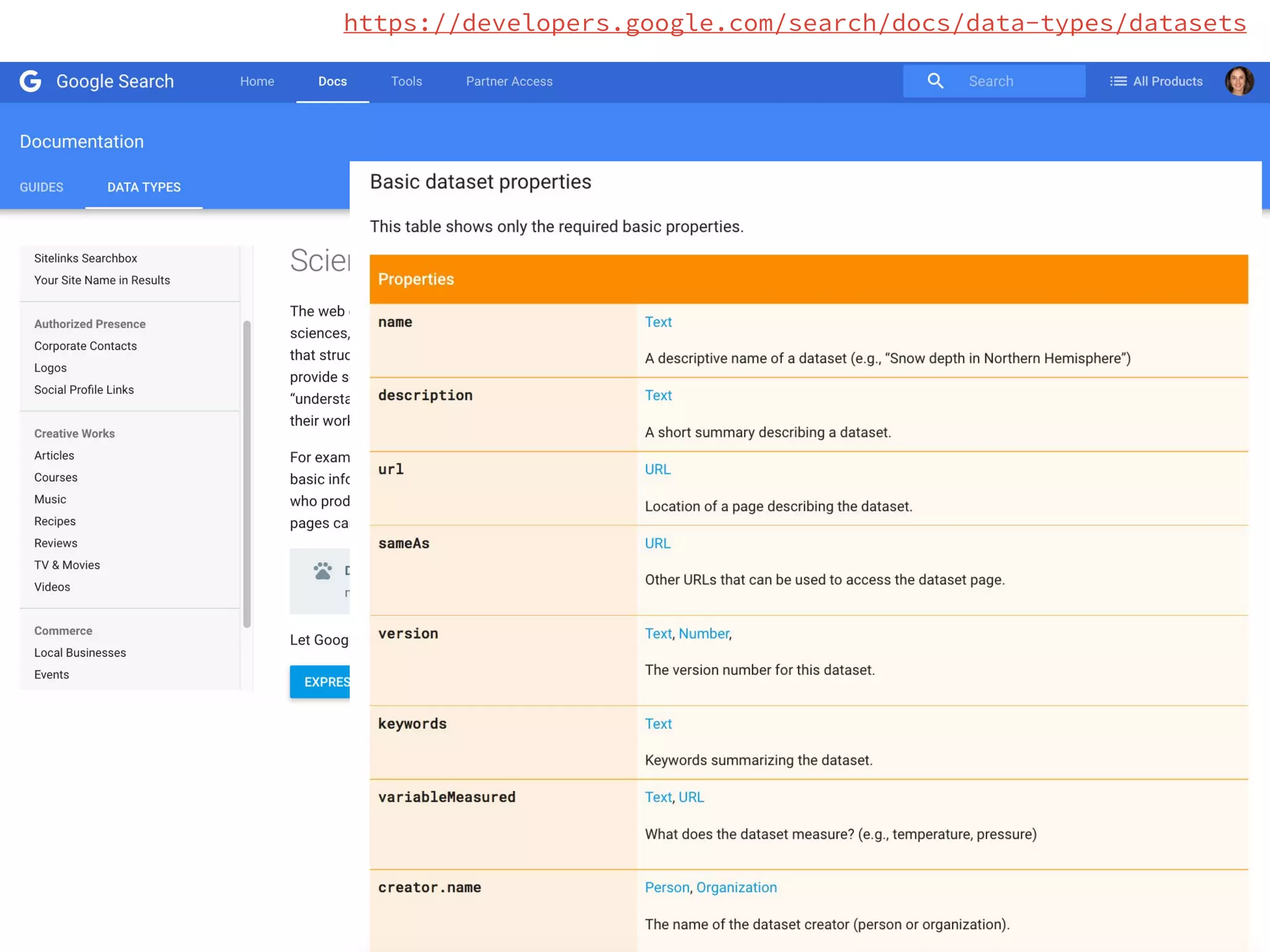
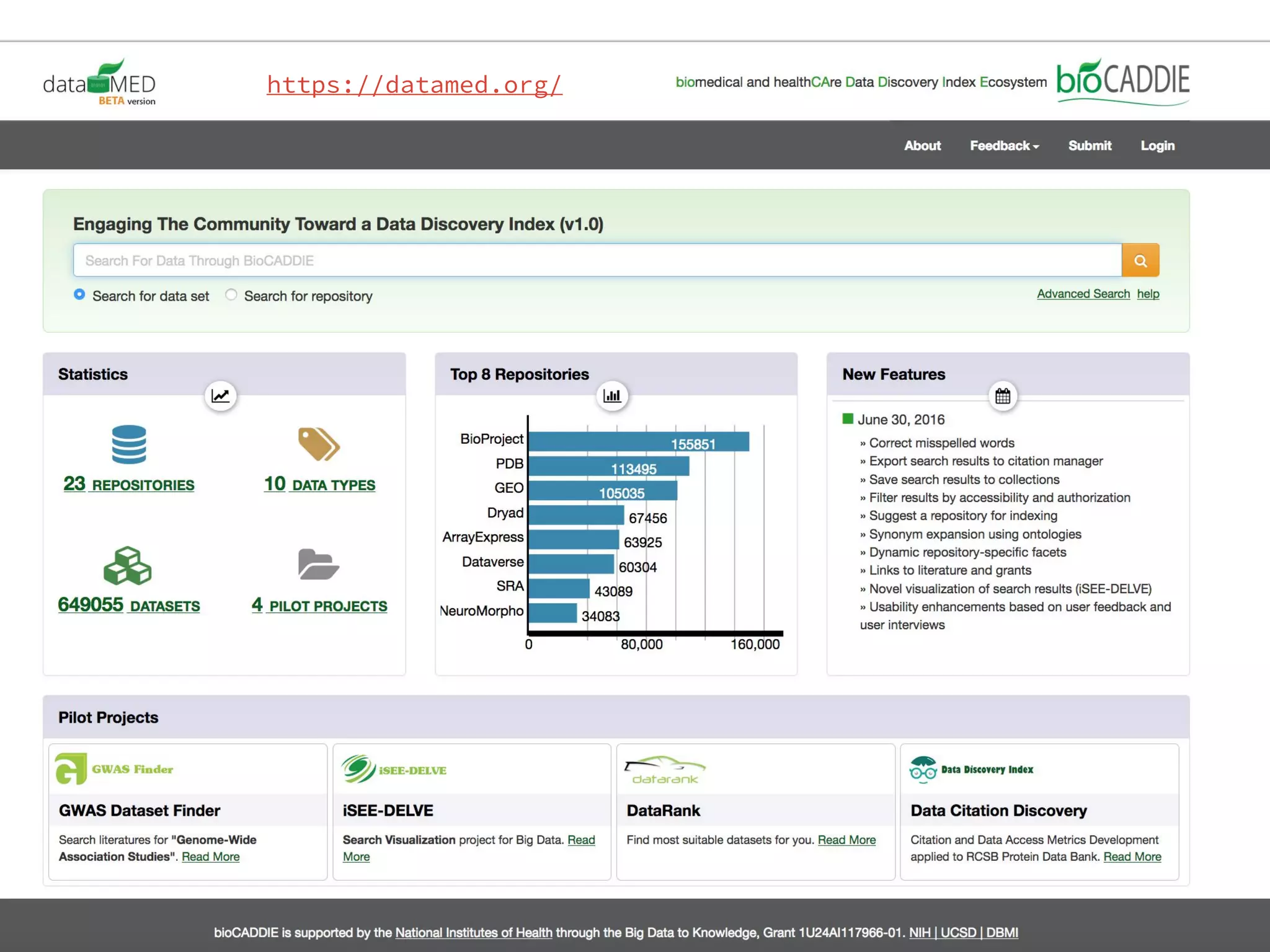
- Efforts are underway to map DATS to schema.org and integrate it within initiatives like DataMed to facilitate indexing of datasets.
- Learning from the DATS experience, the next steps involve developing a Bioschemas specification focused specifically