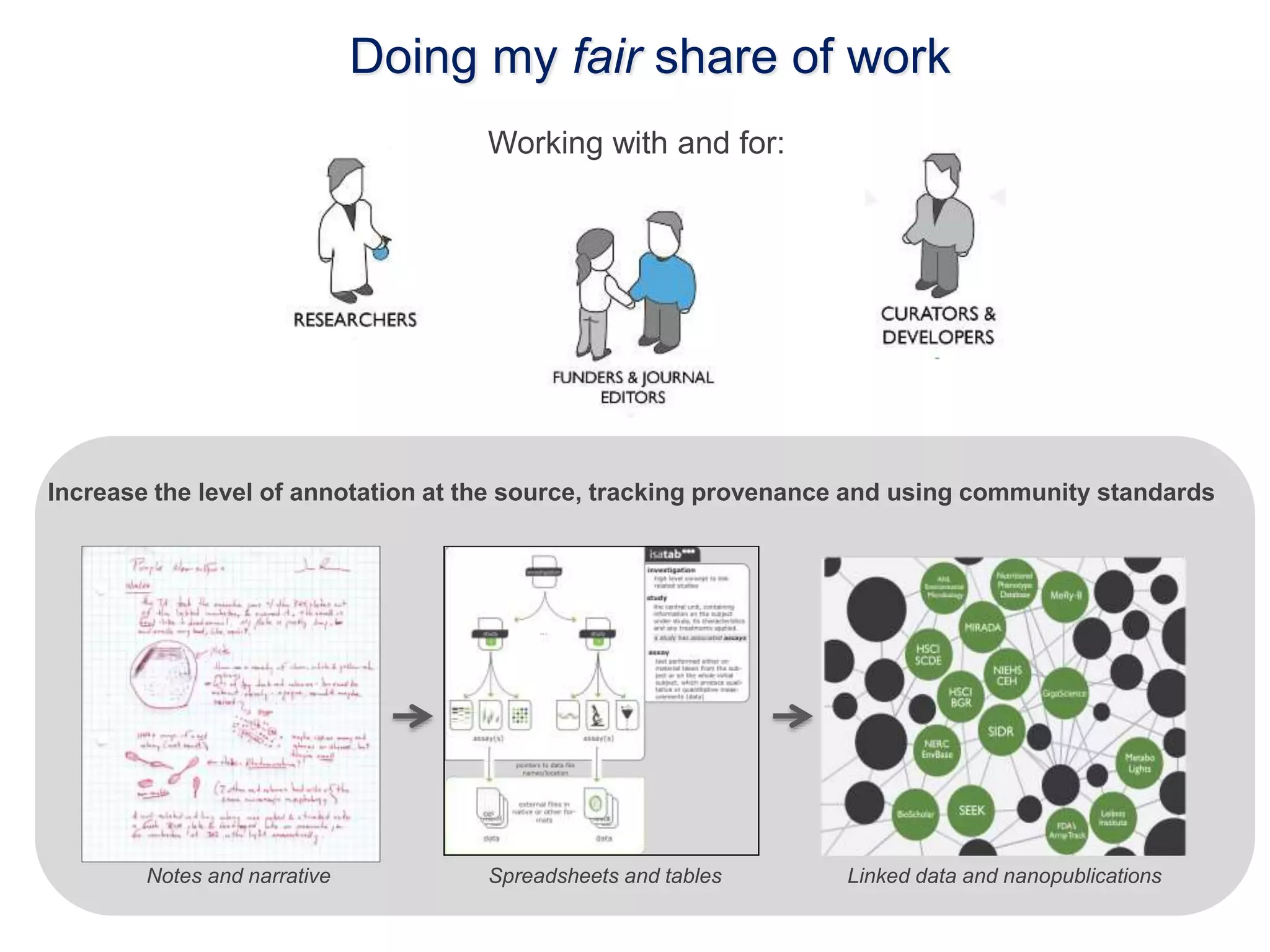
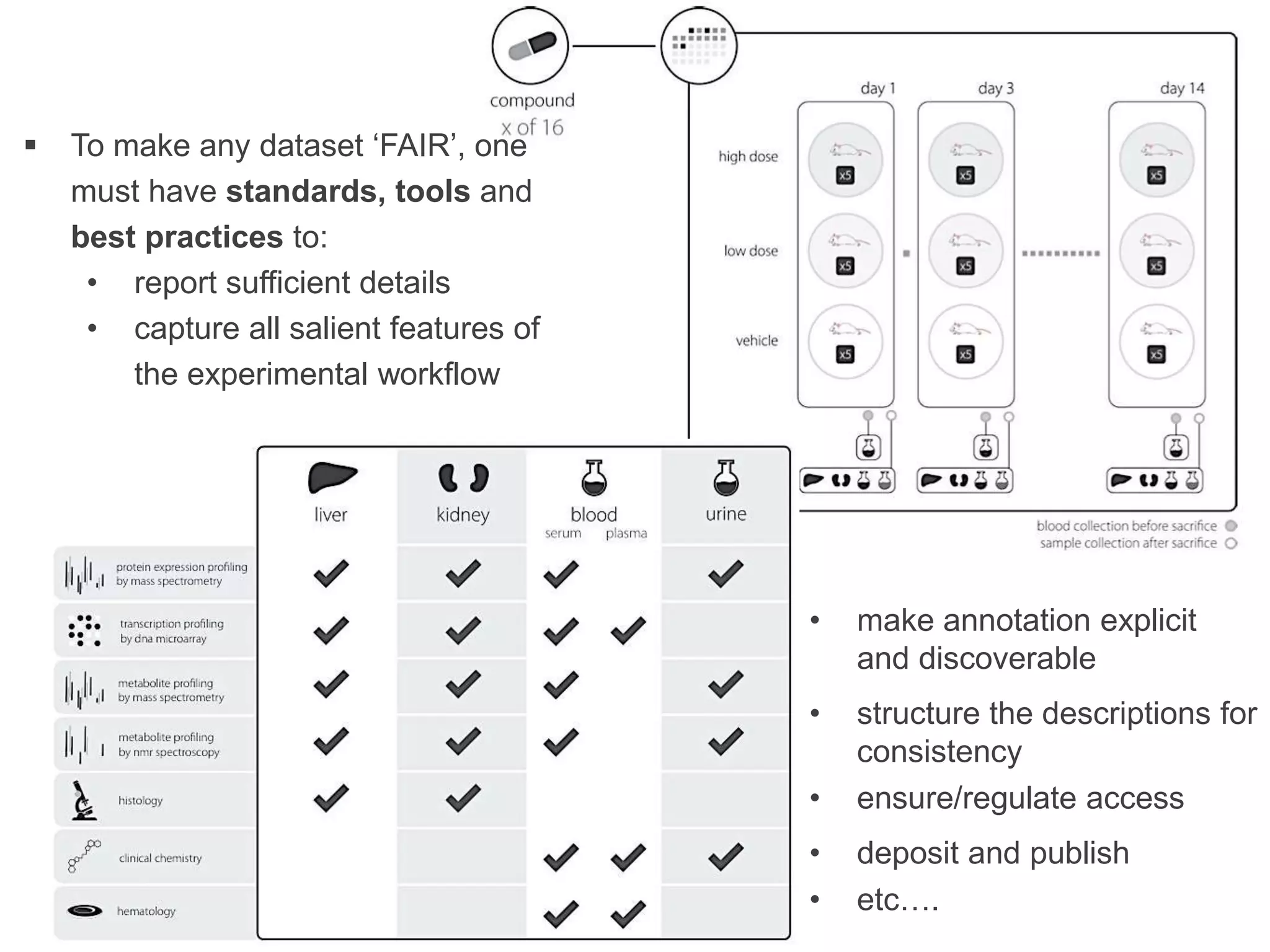
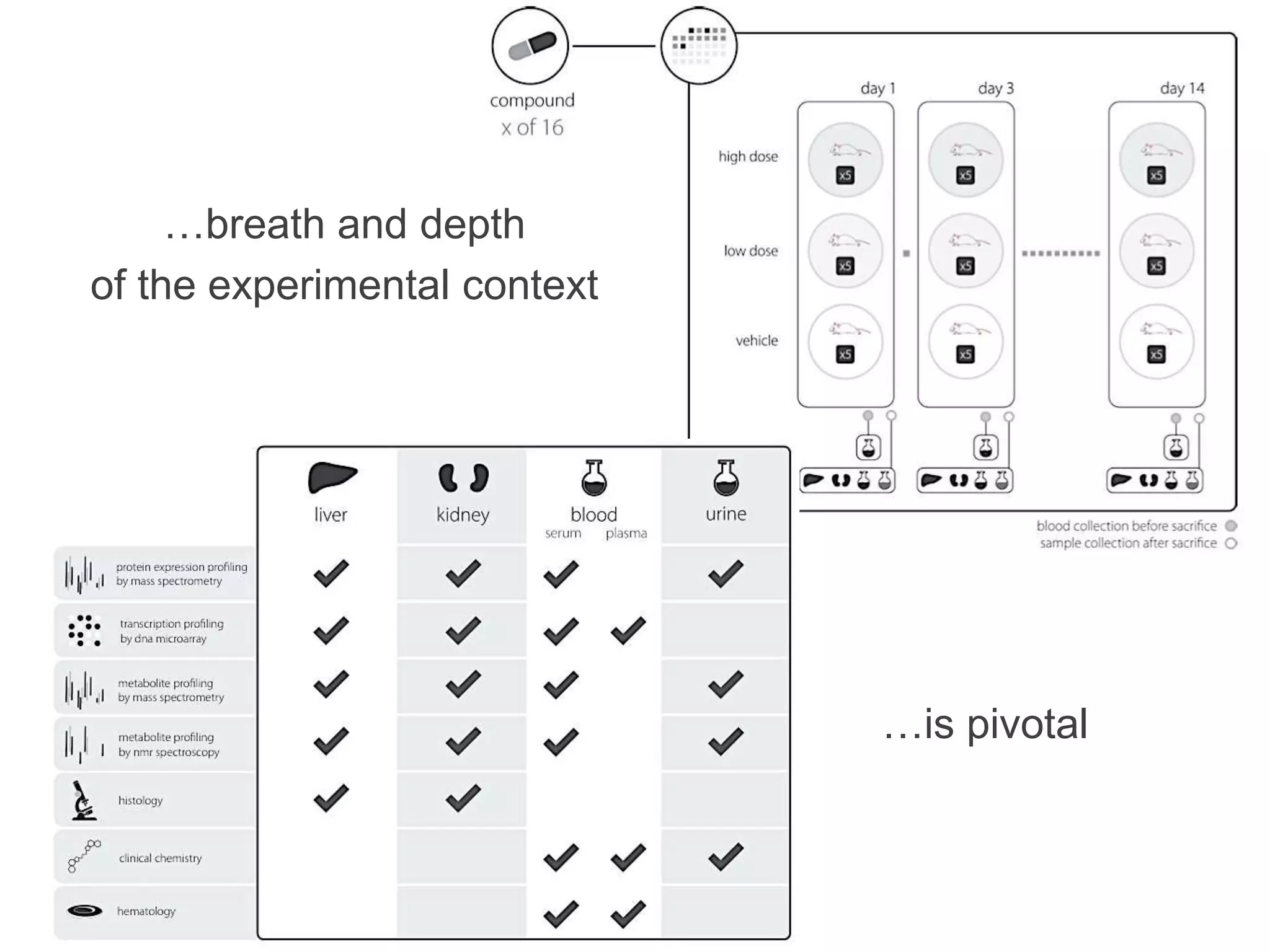
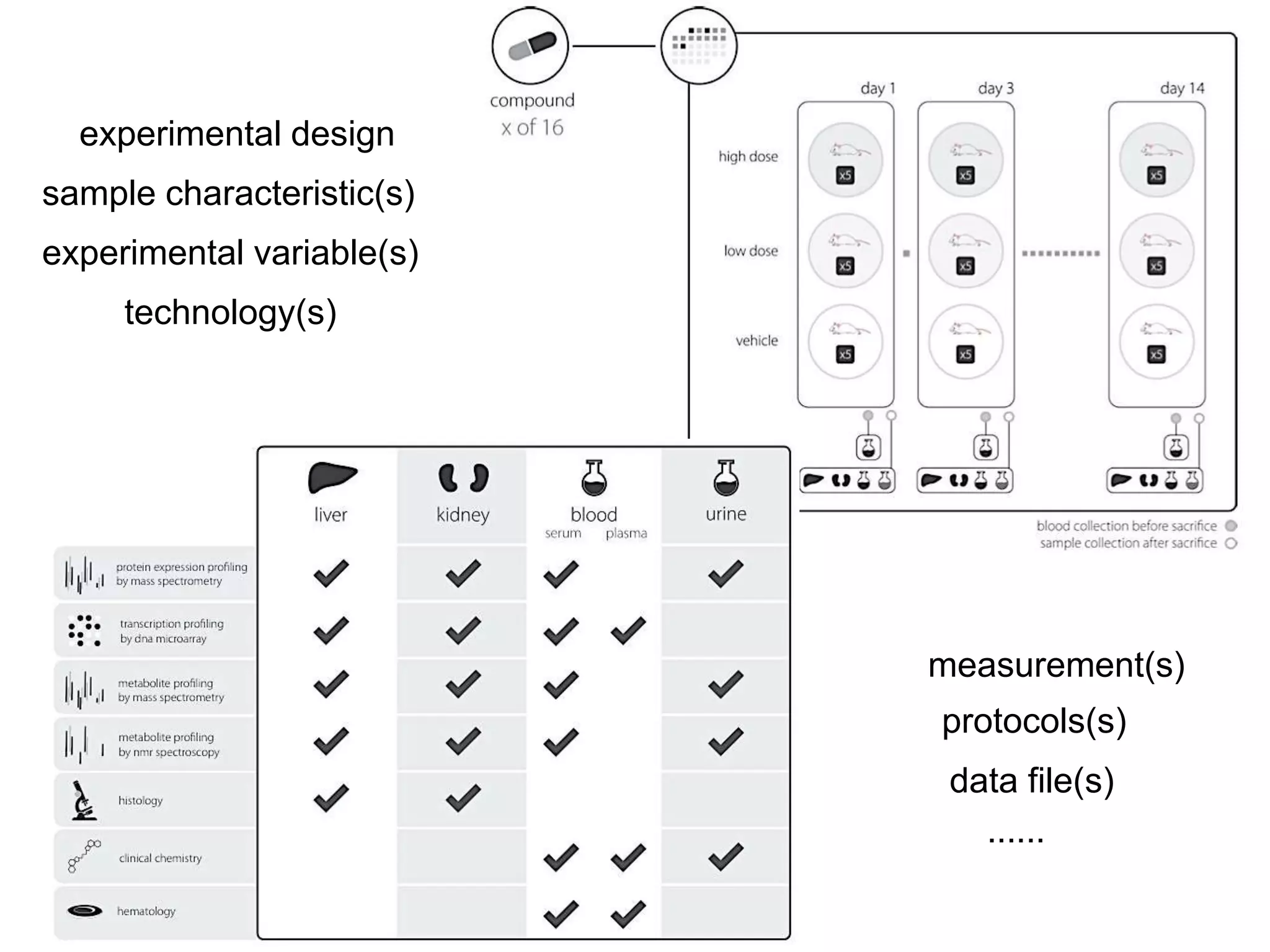
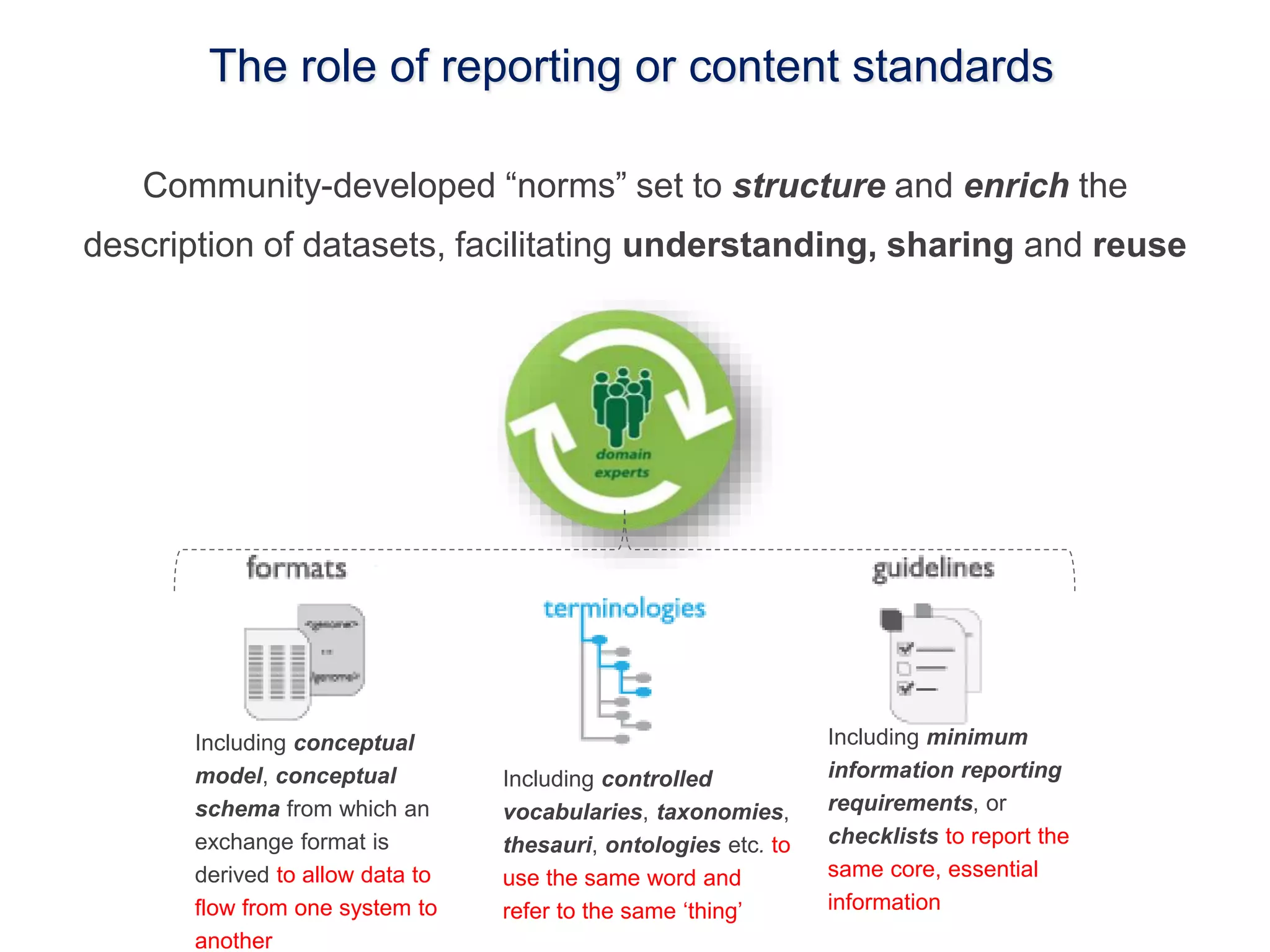
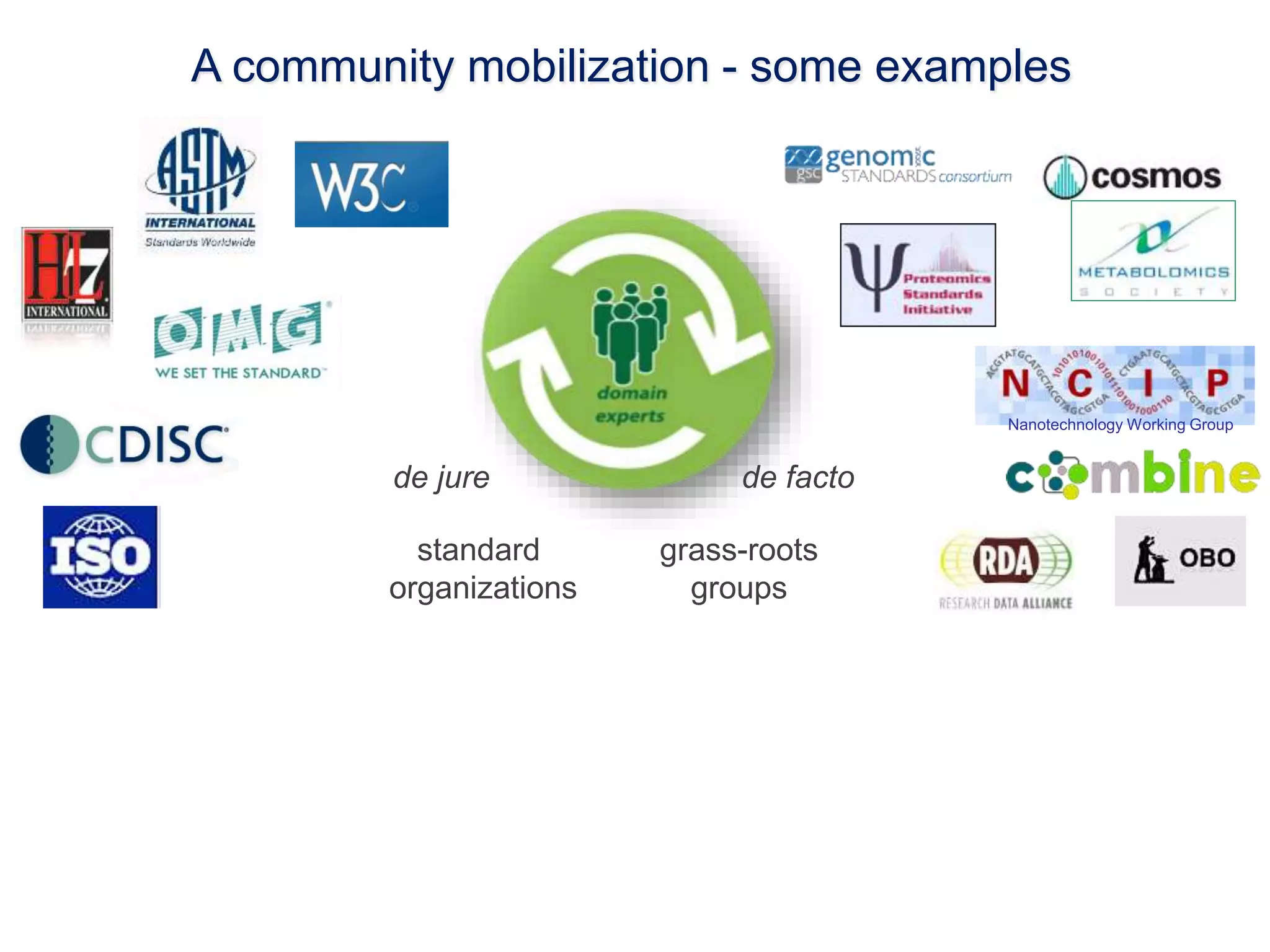
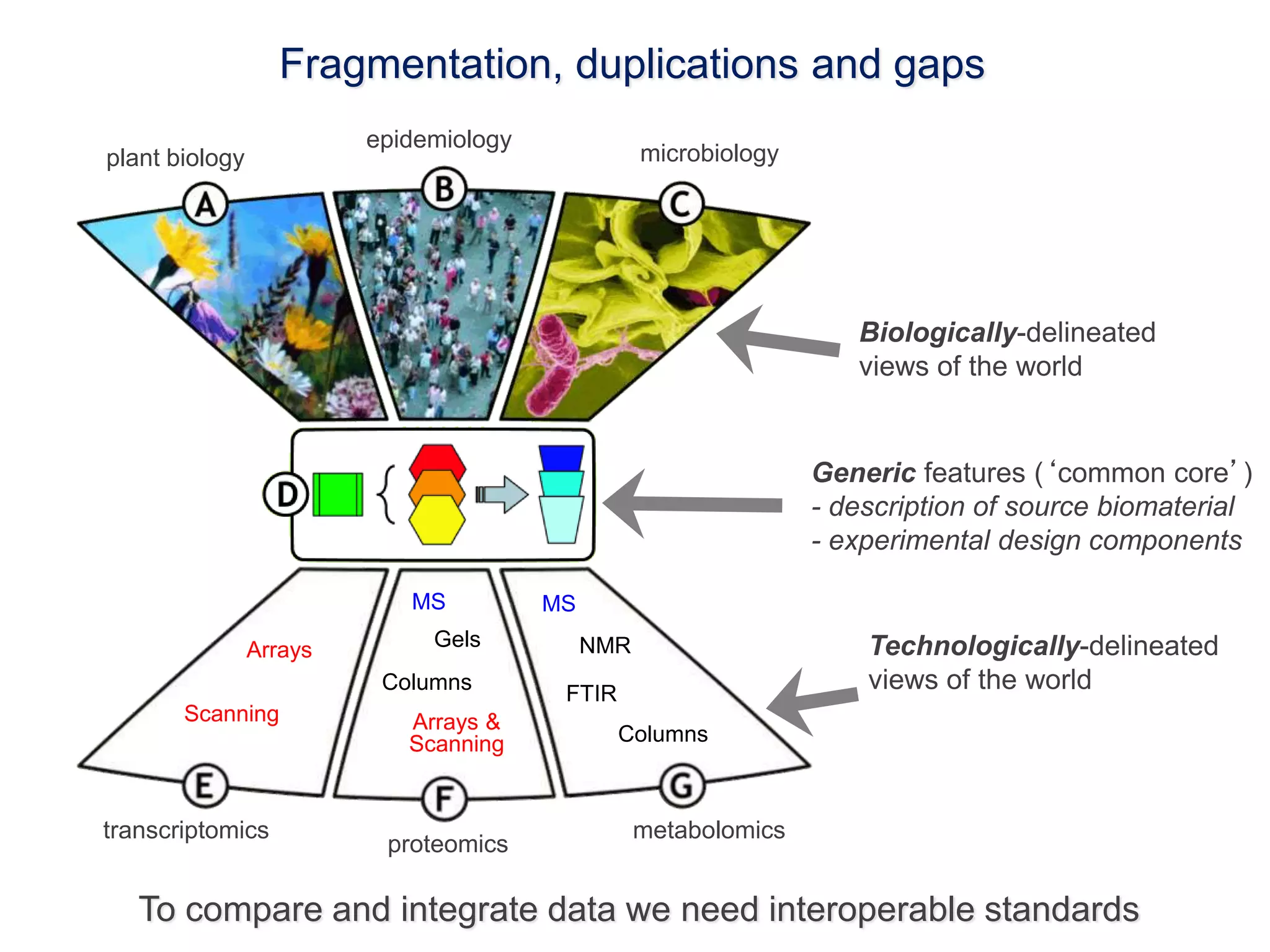
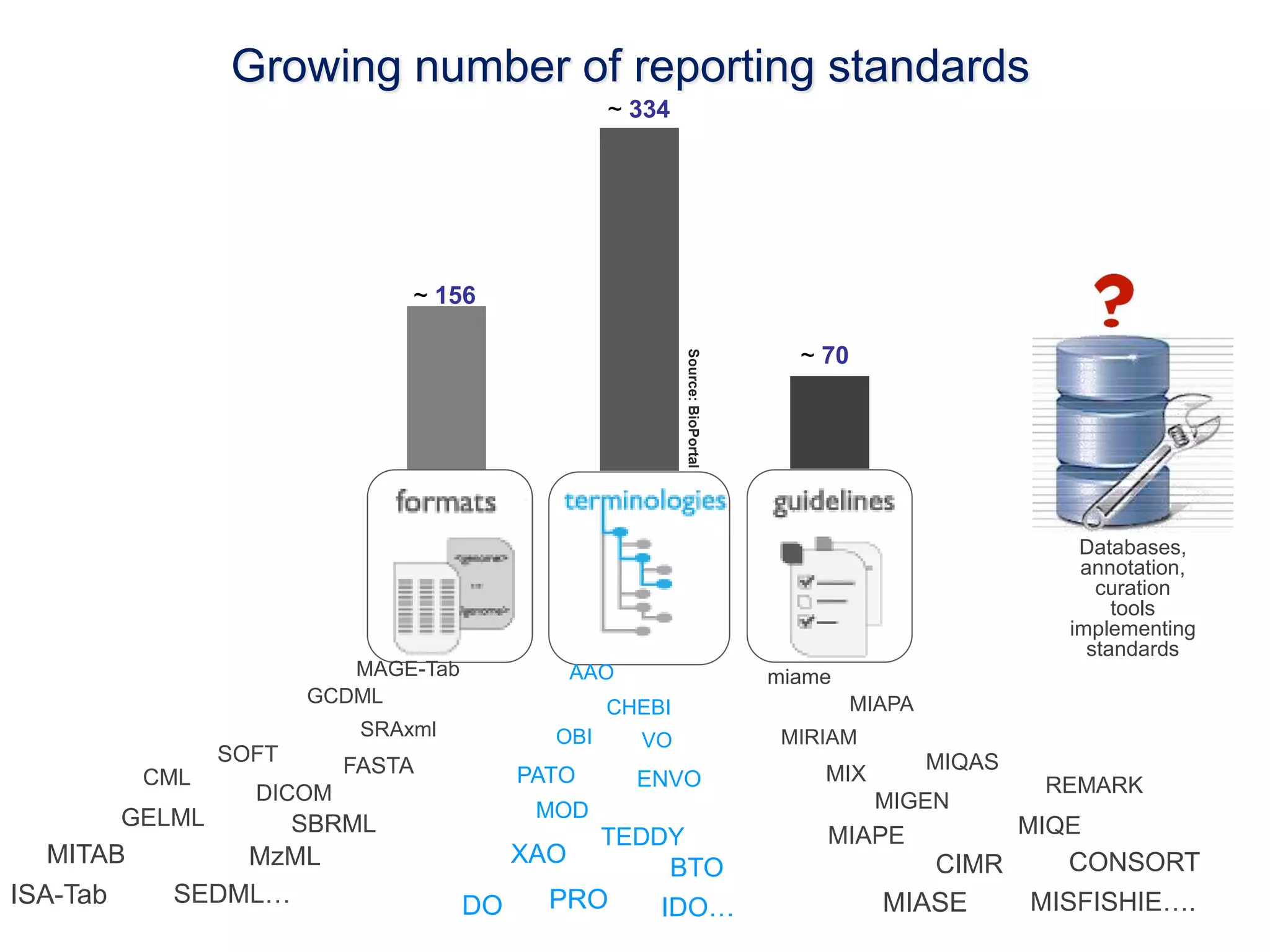
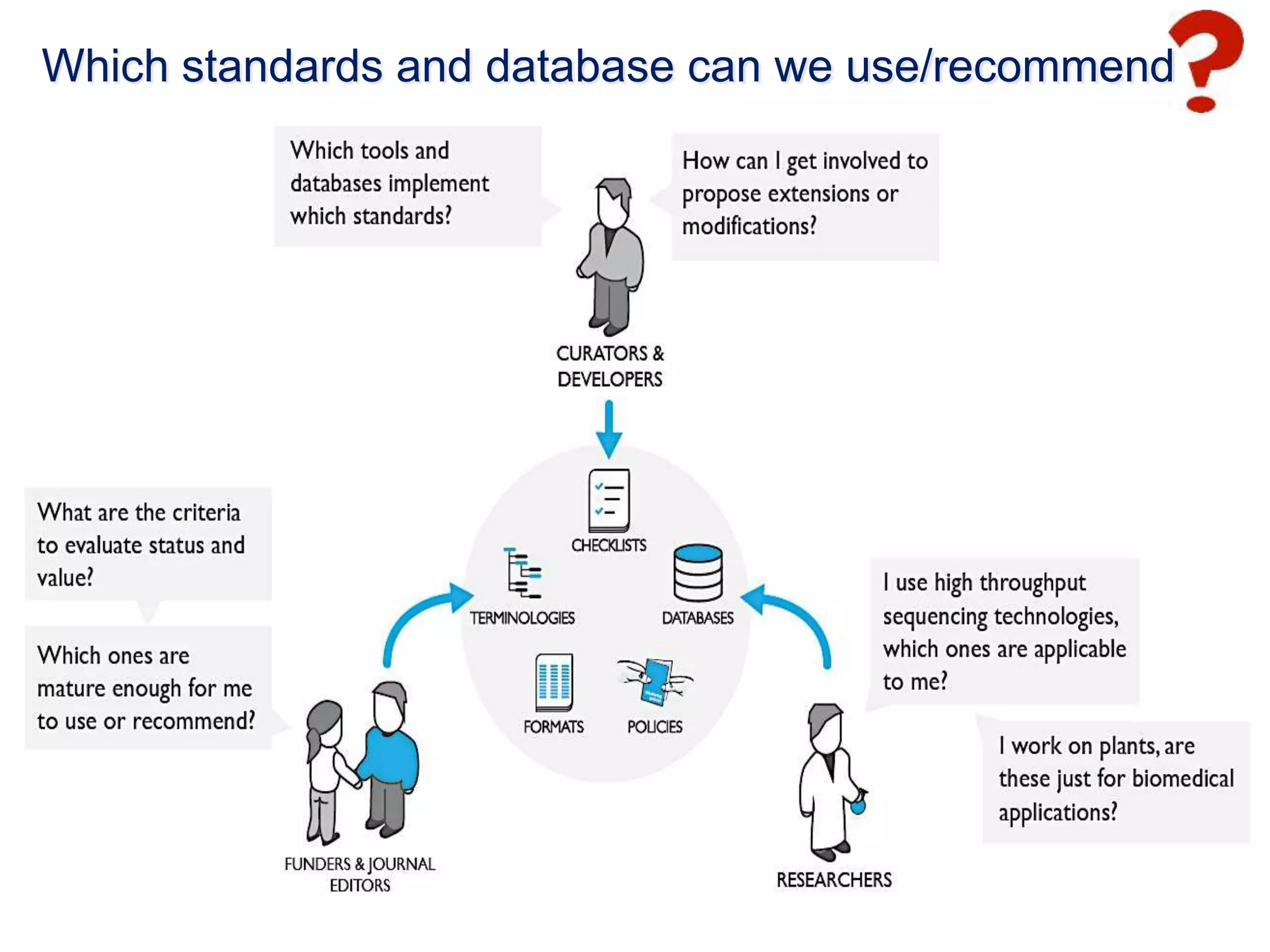
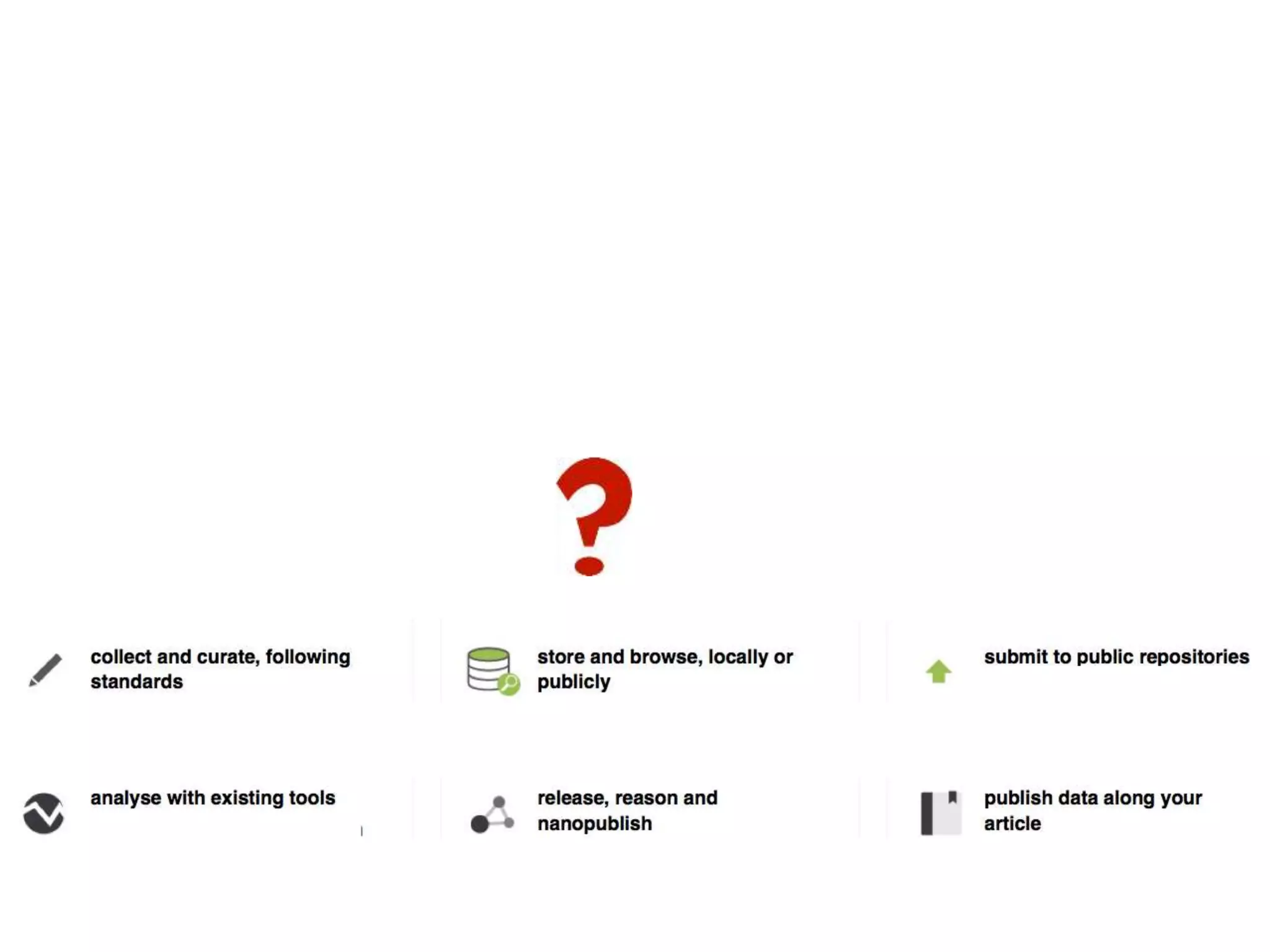
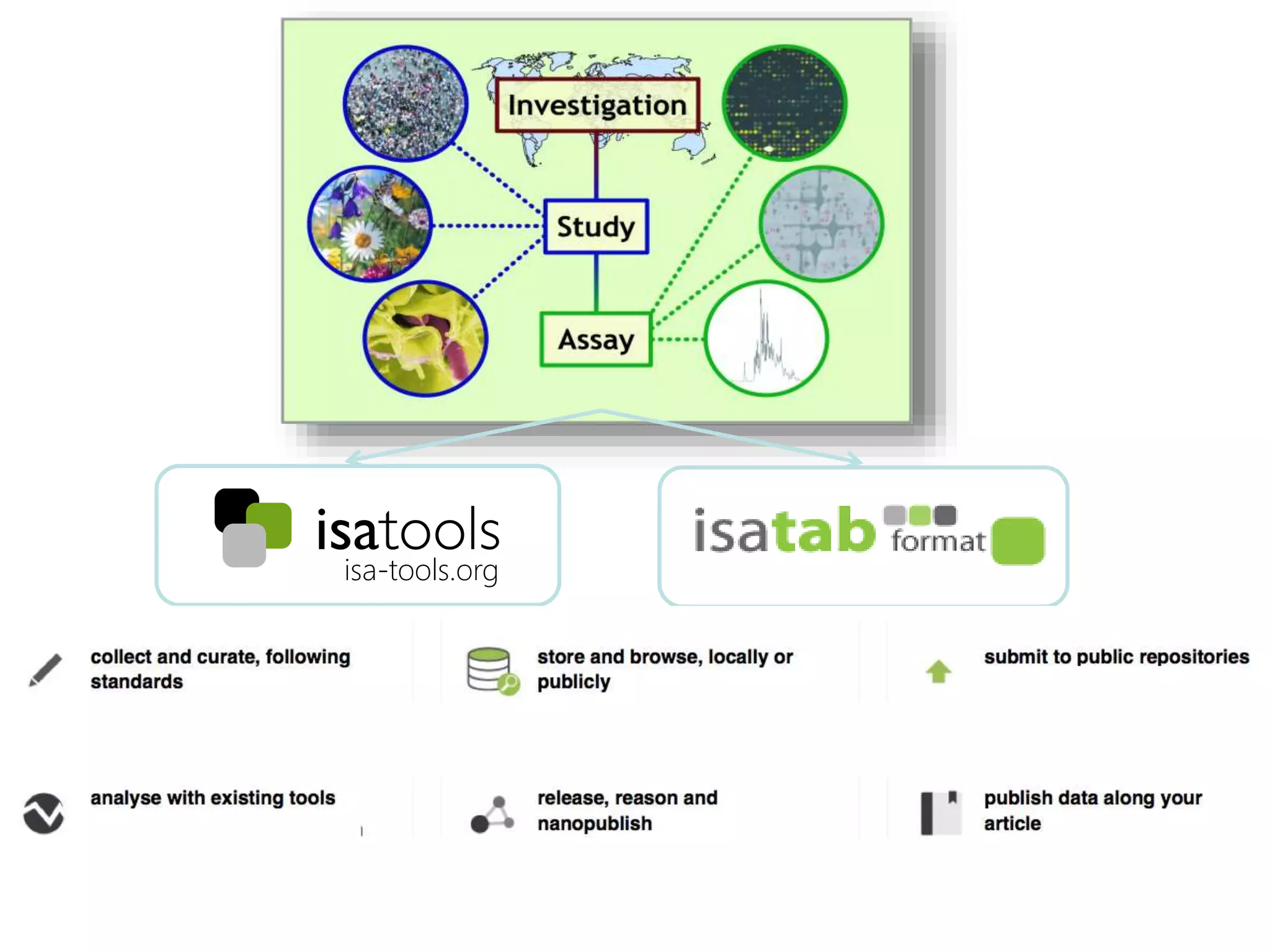
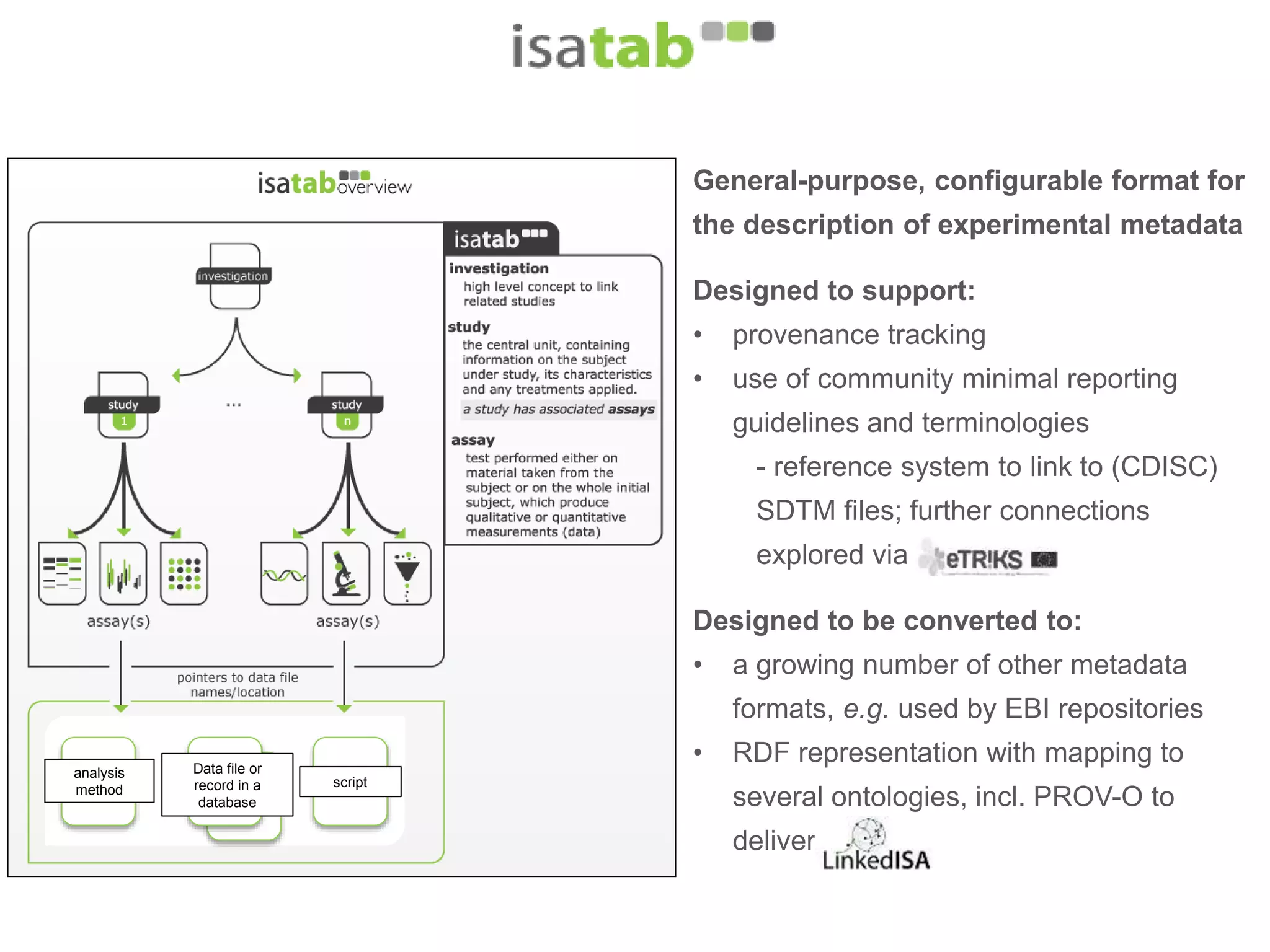
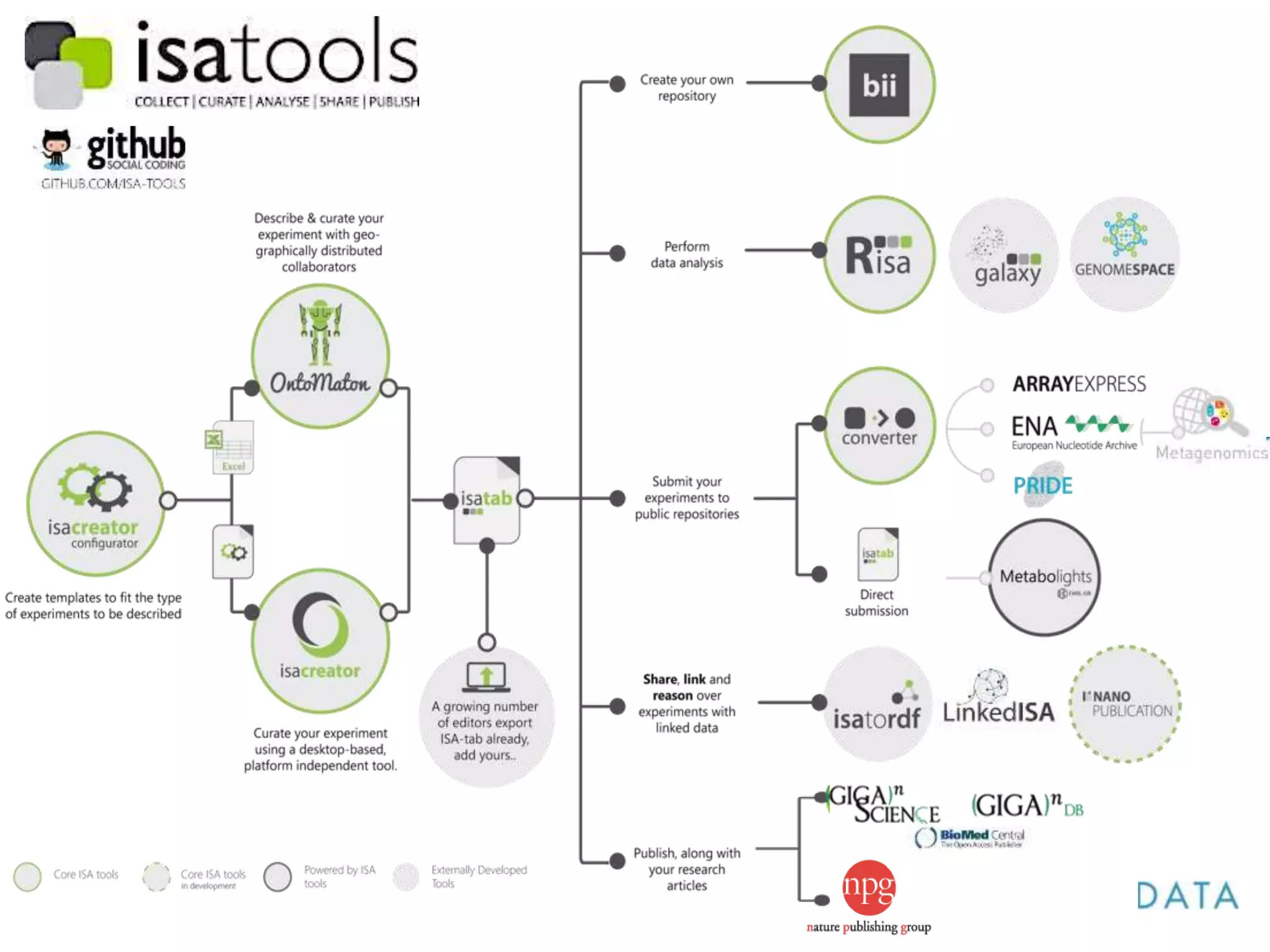
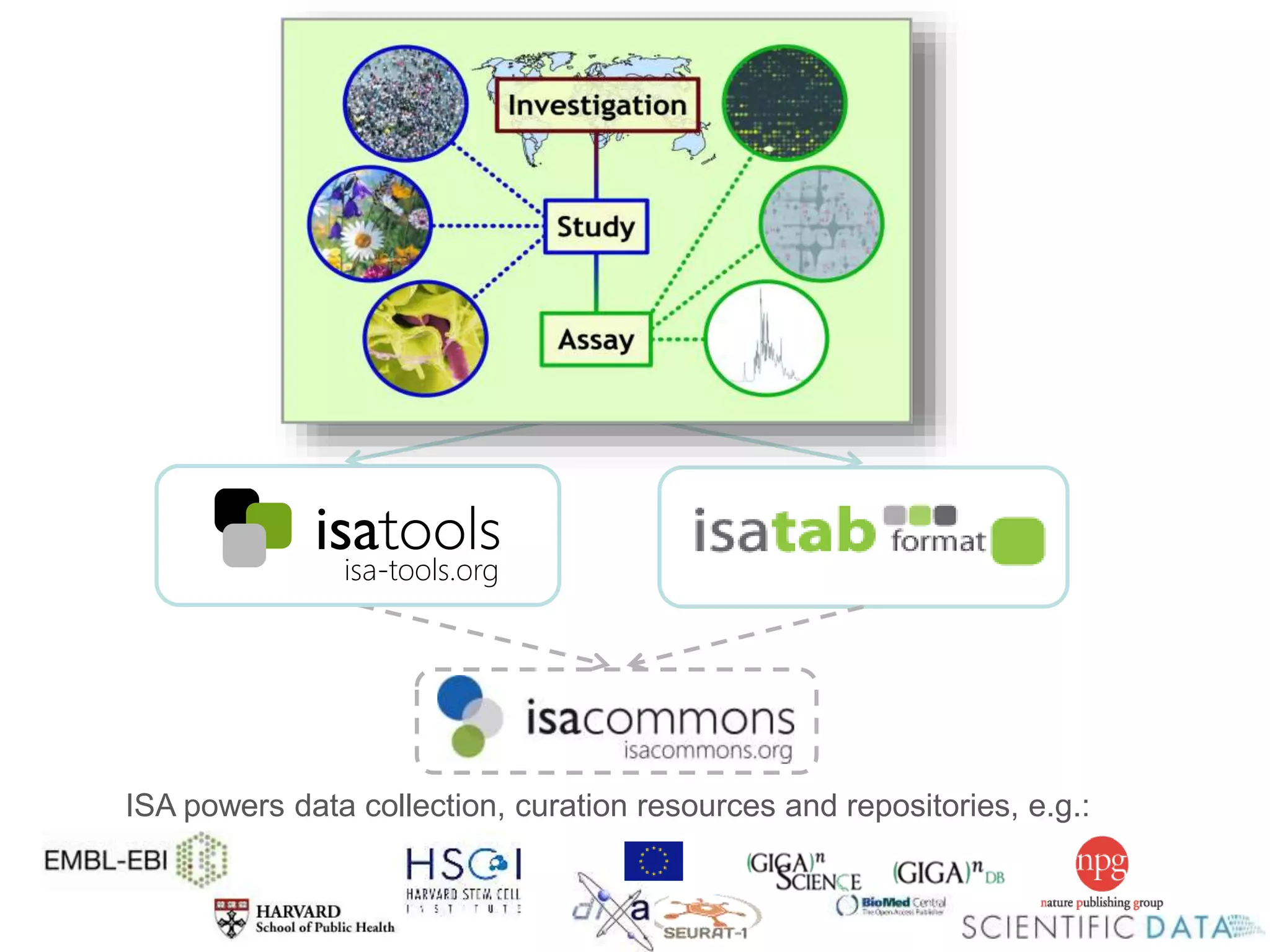
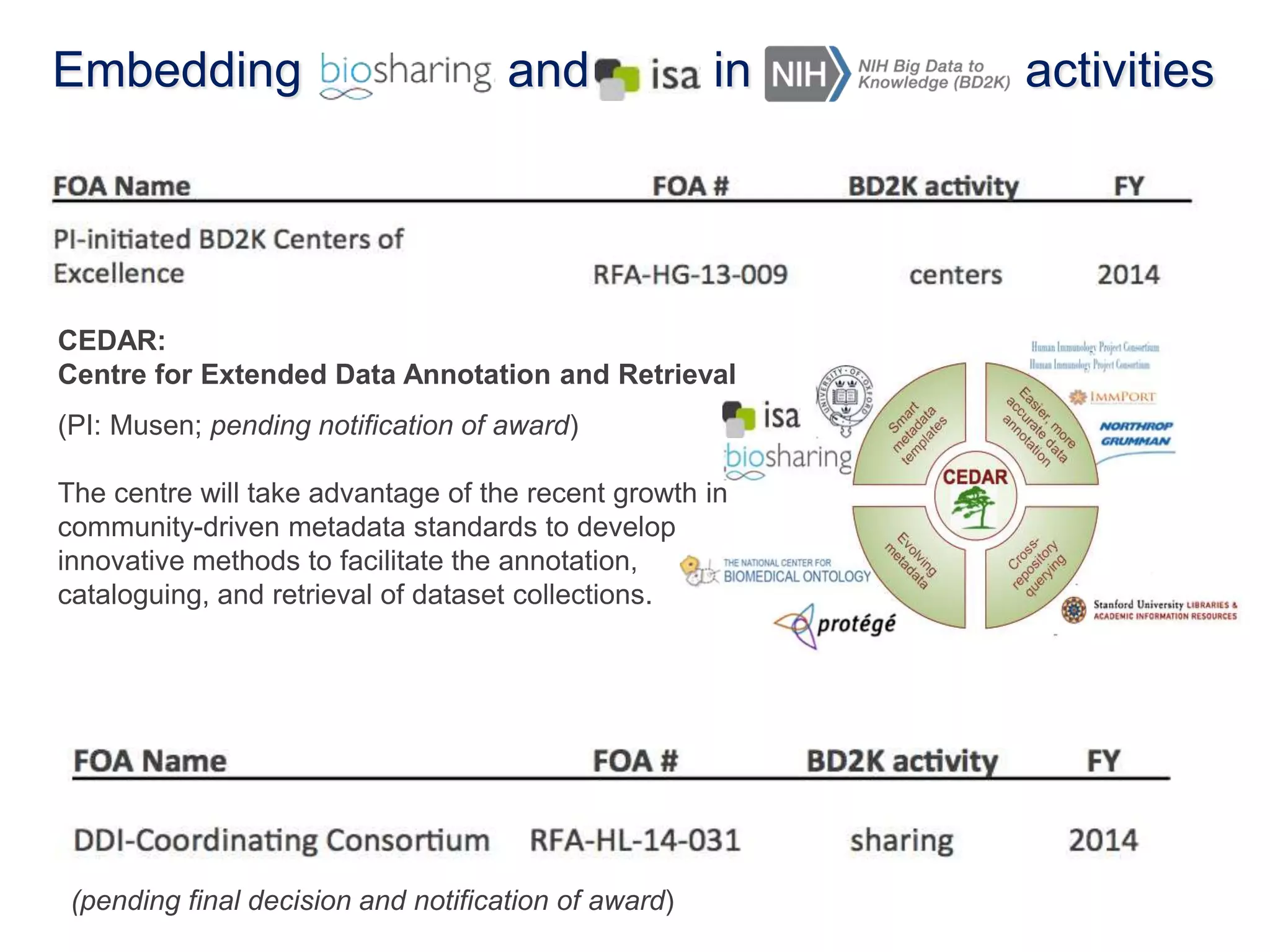
1) The document discusses Susanna-Assunta Sansone's roles and work related to promoting FAIR data standards and practices.
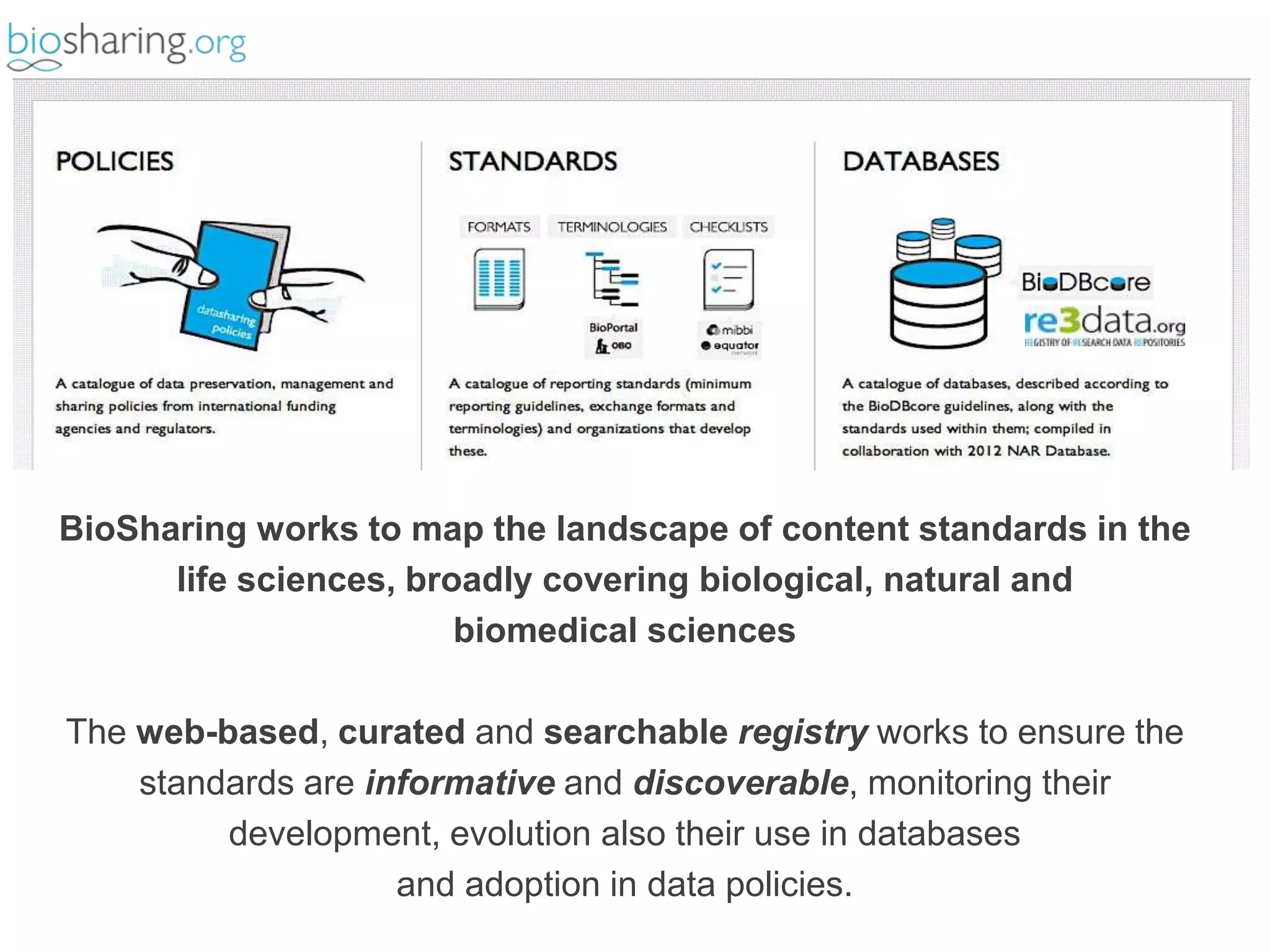
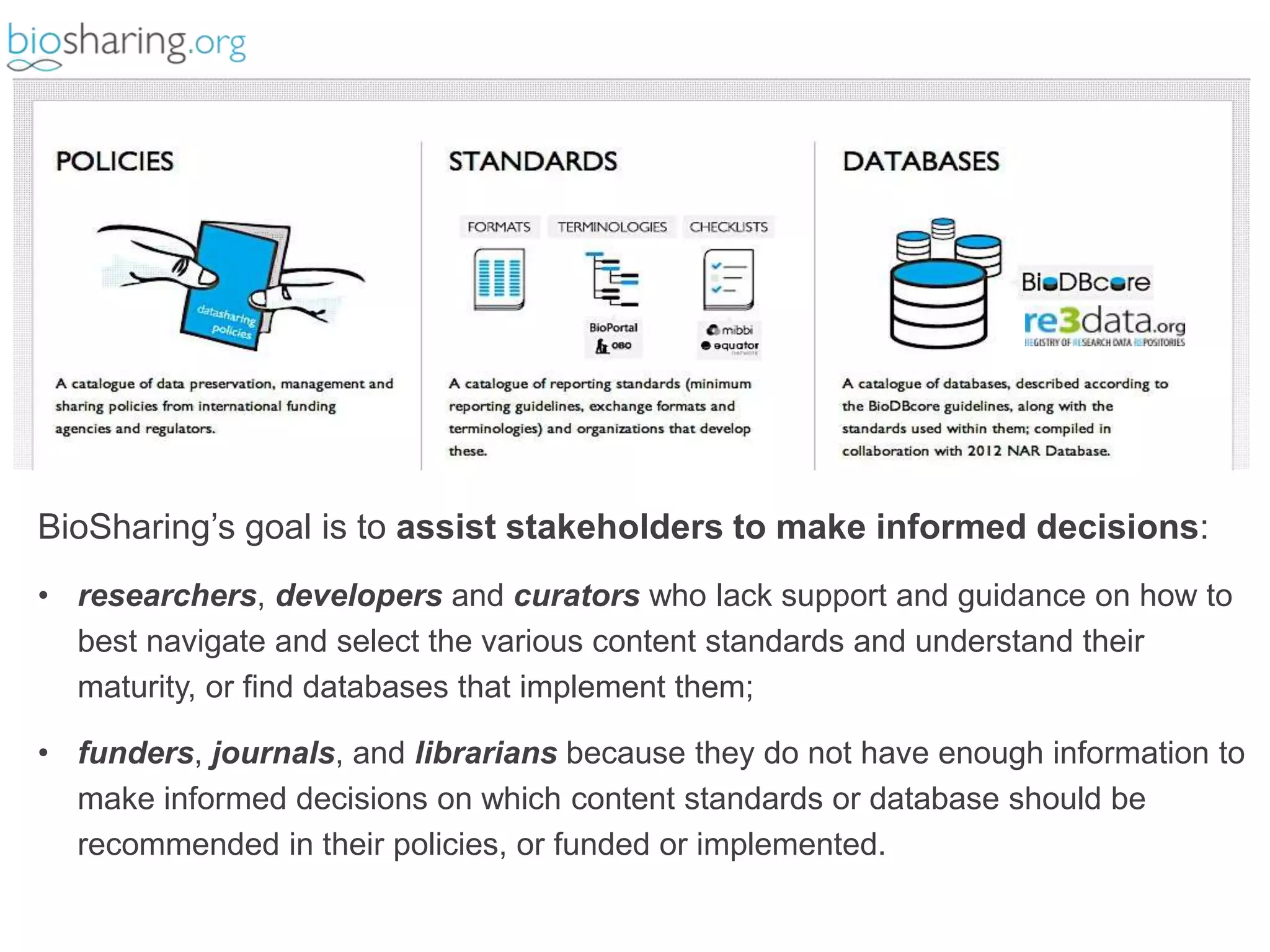
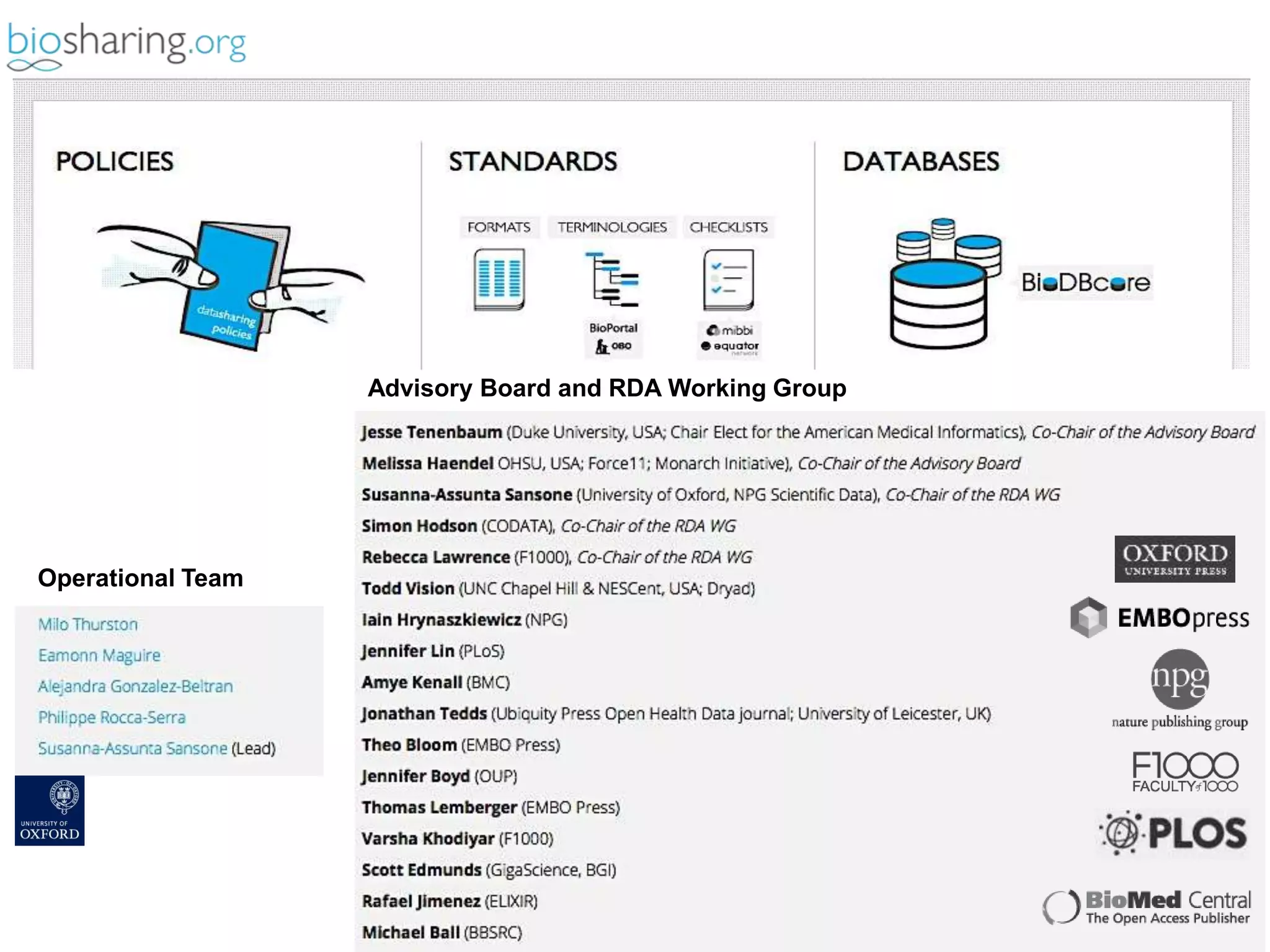
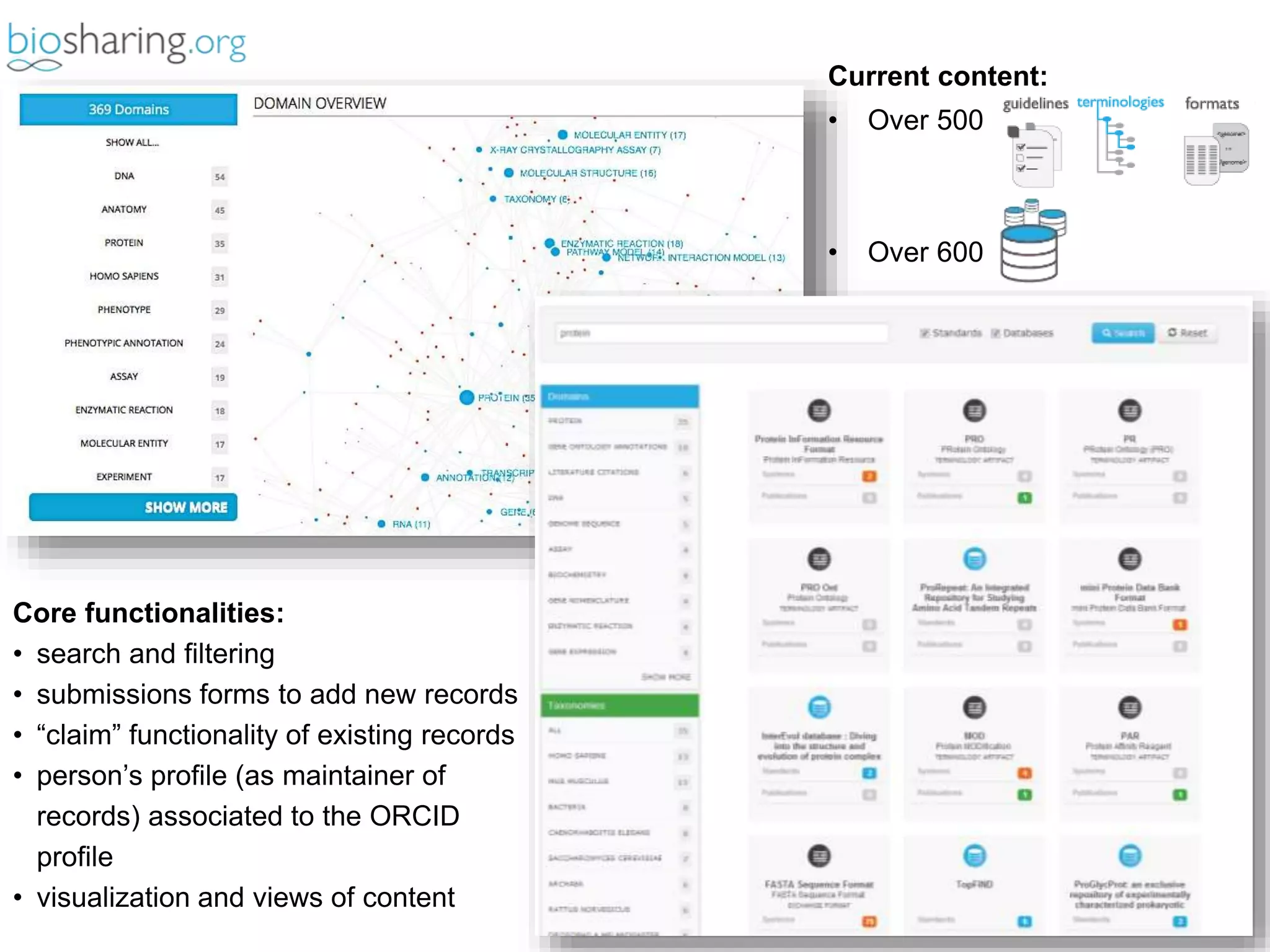
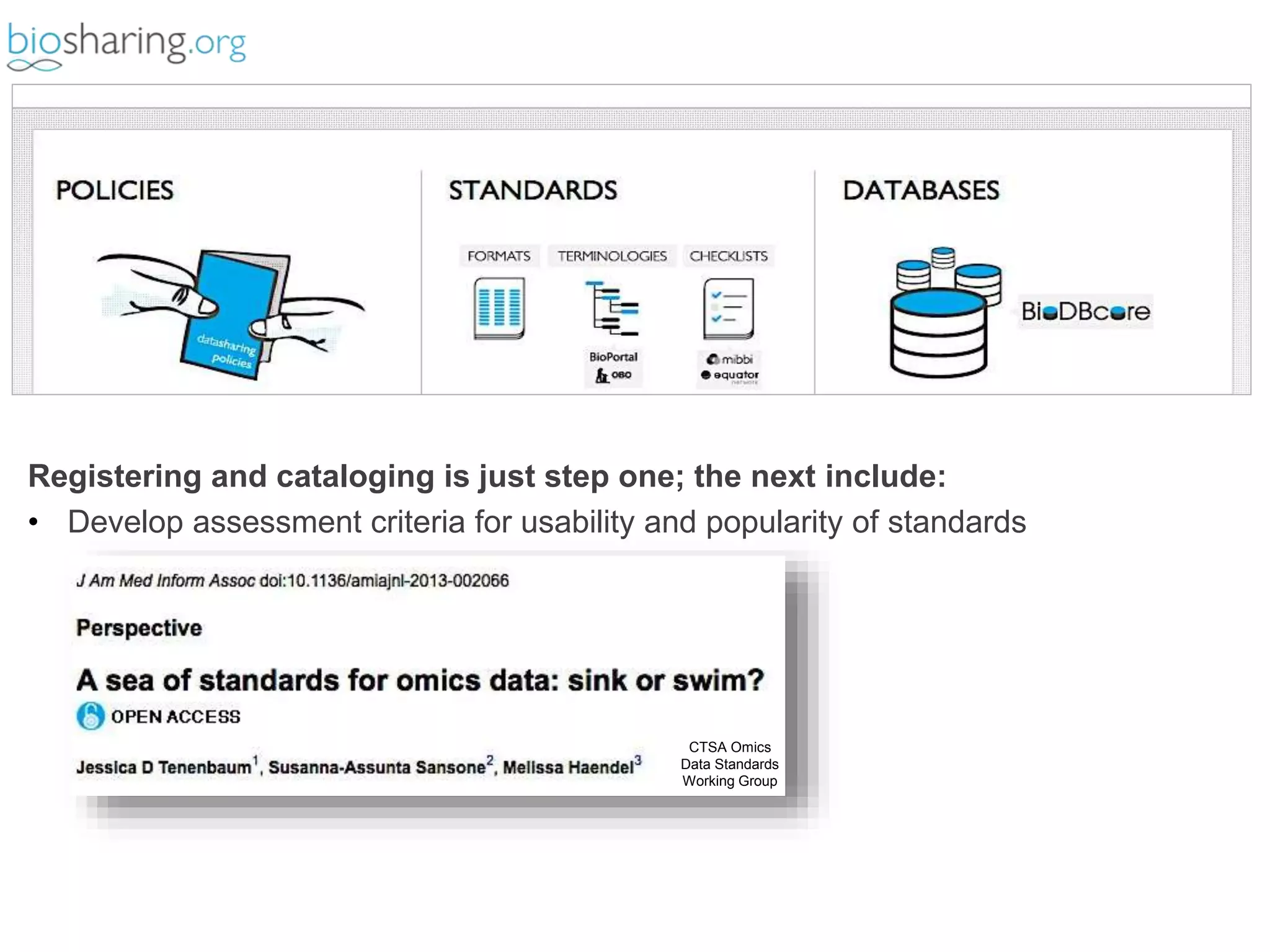
2) It highlights some of her leadership positions with organizations like BioSharing that work to map and promote standards.
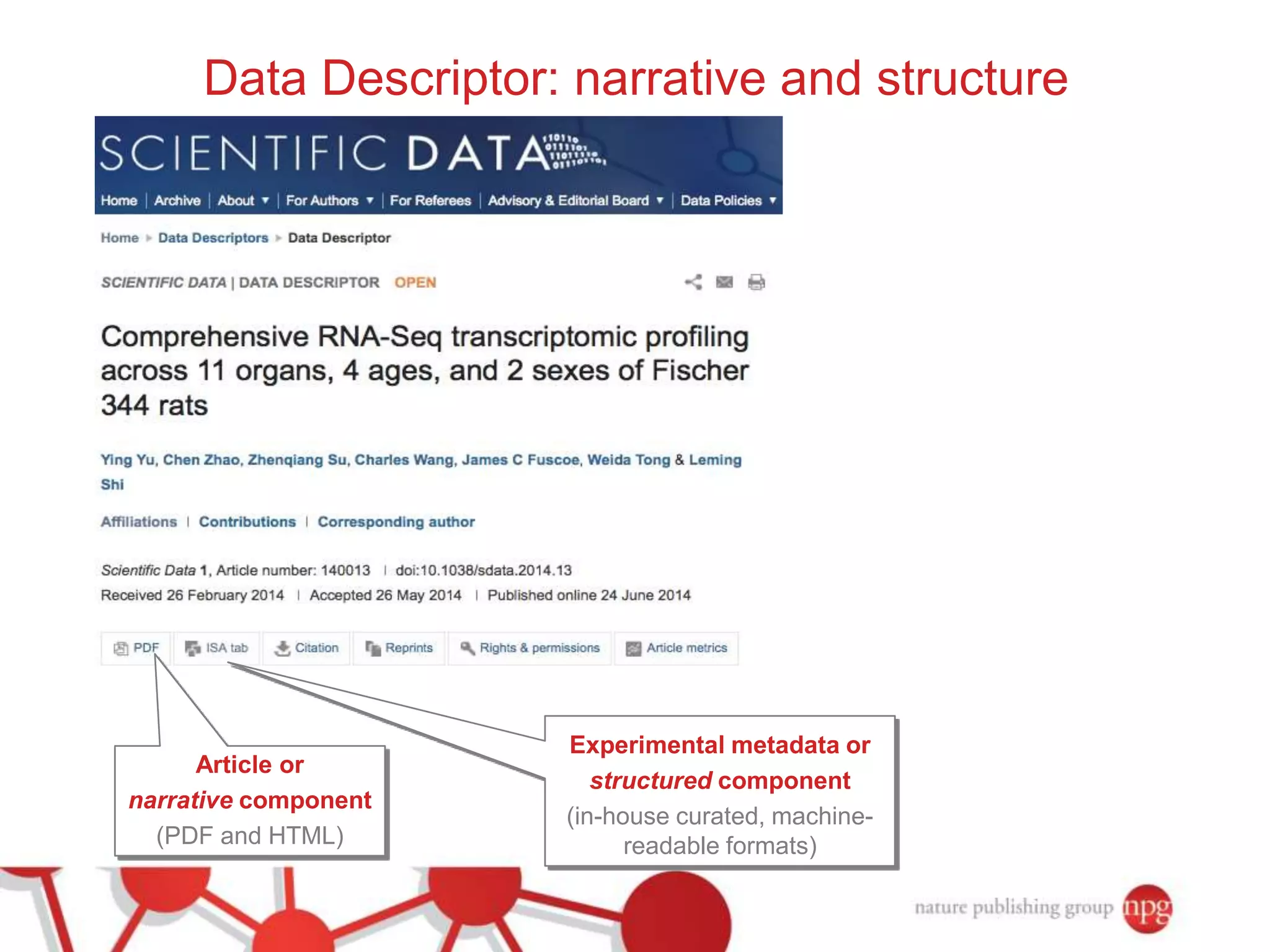
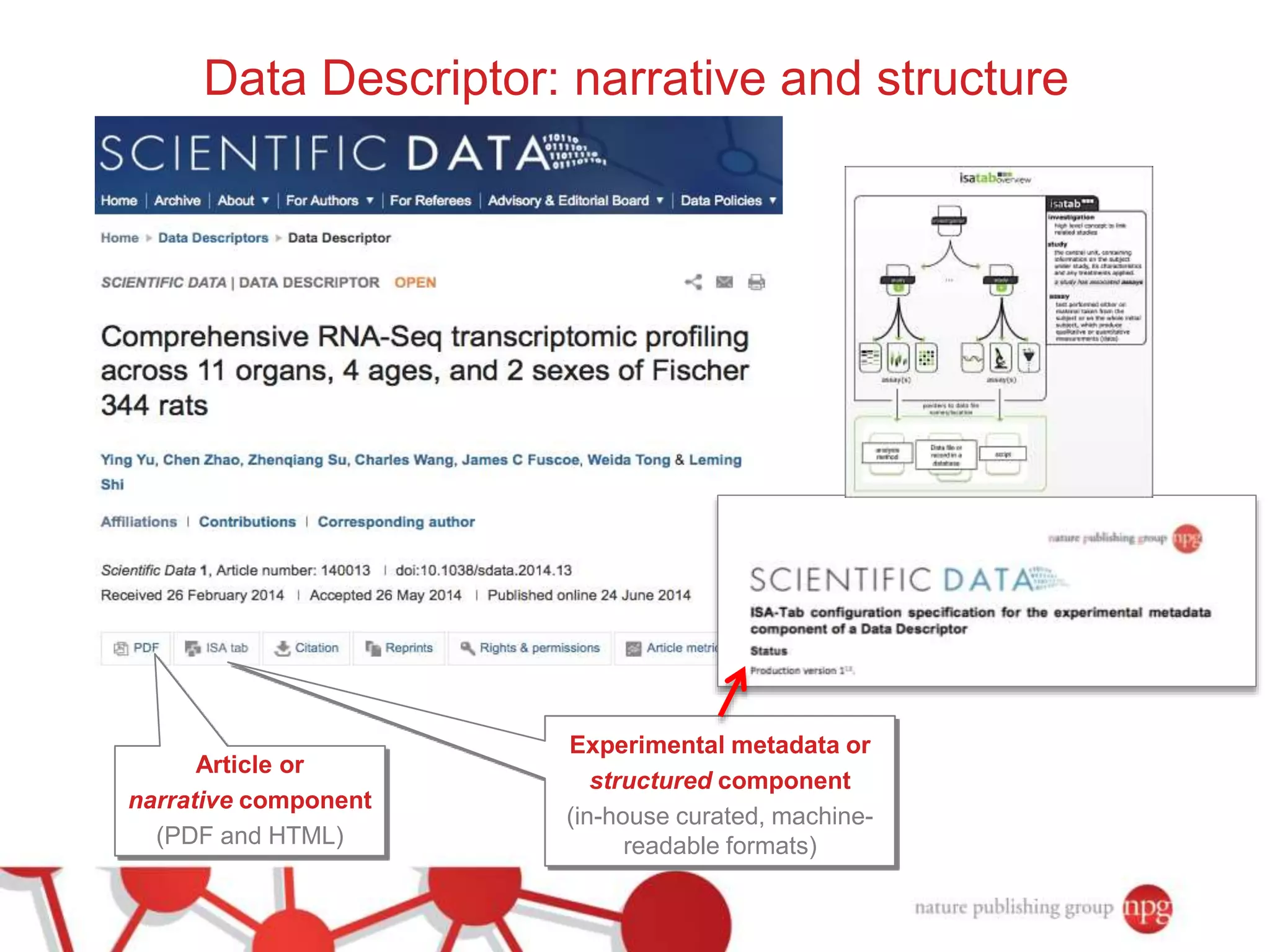
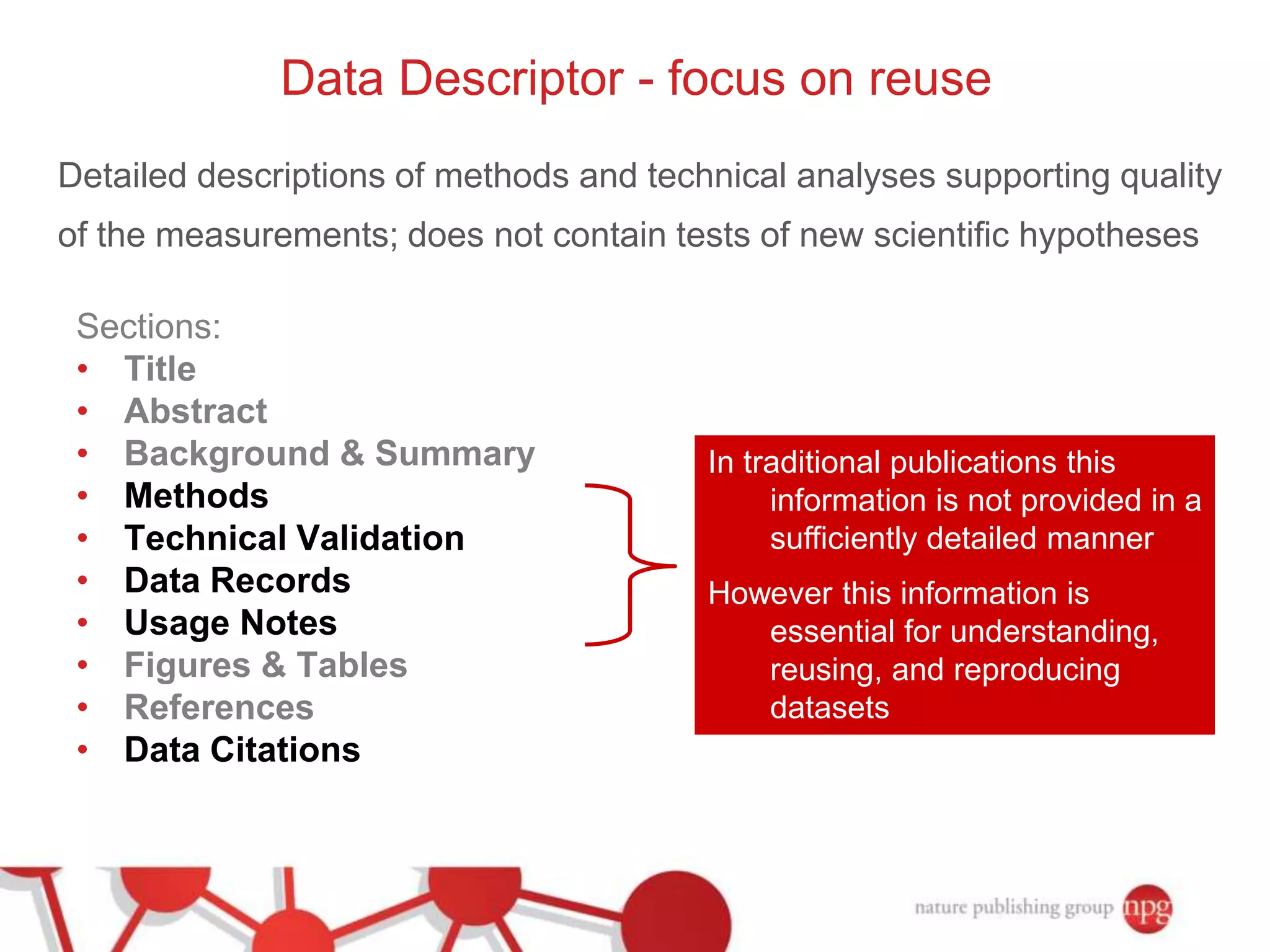
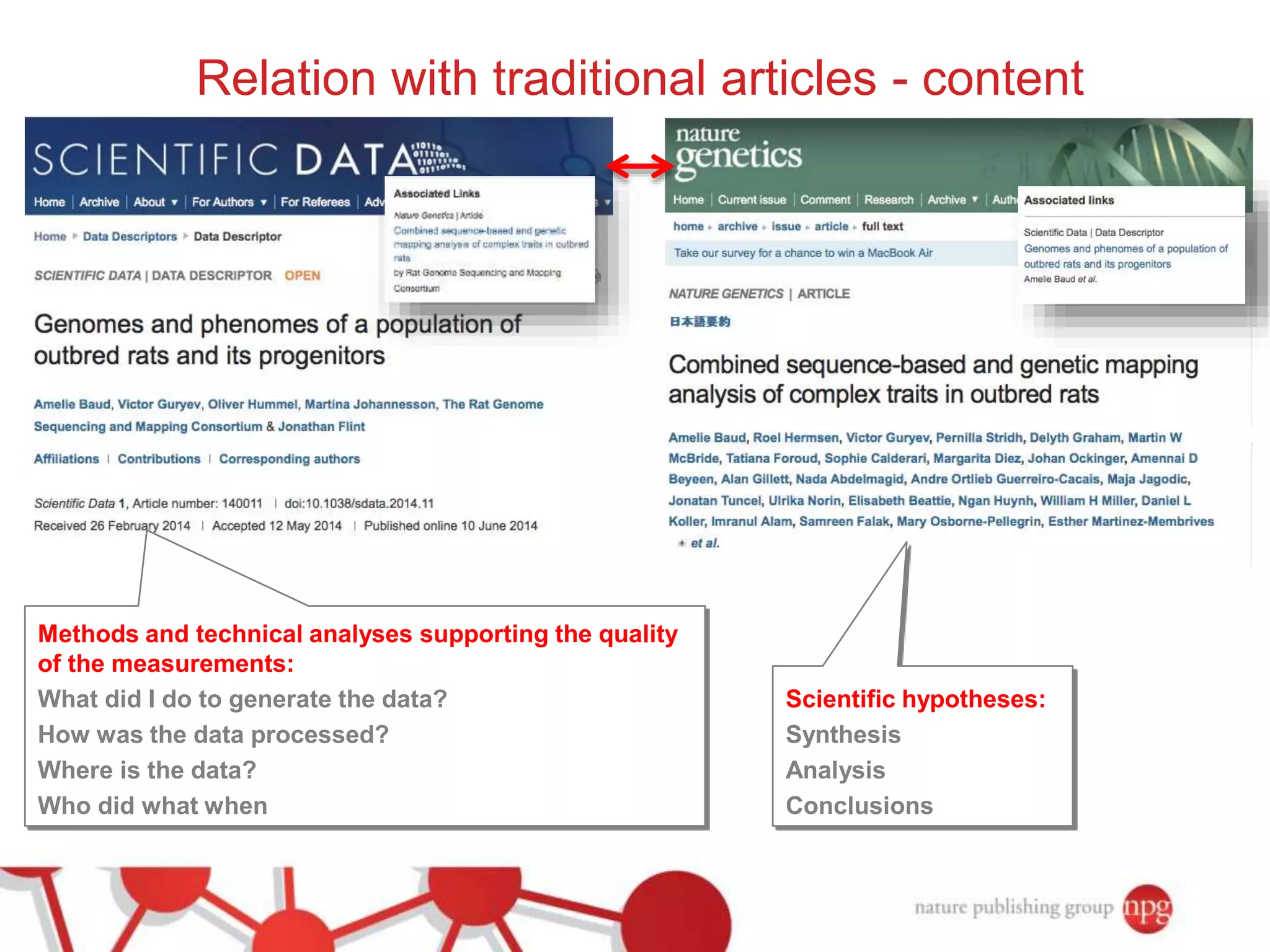
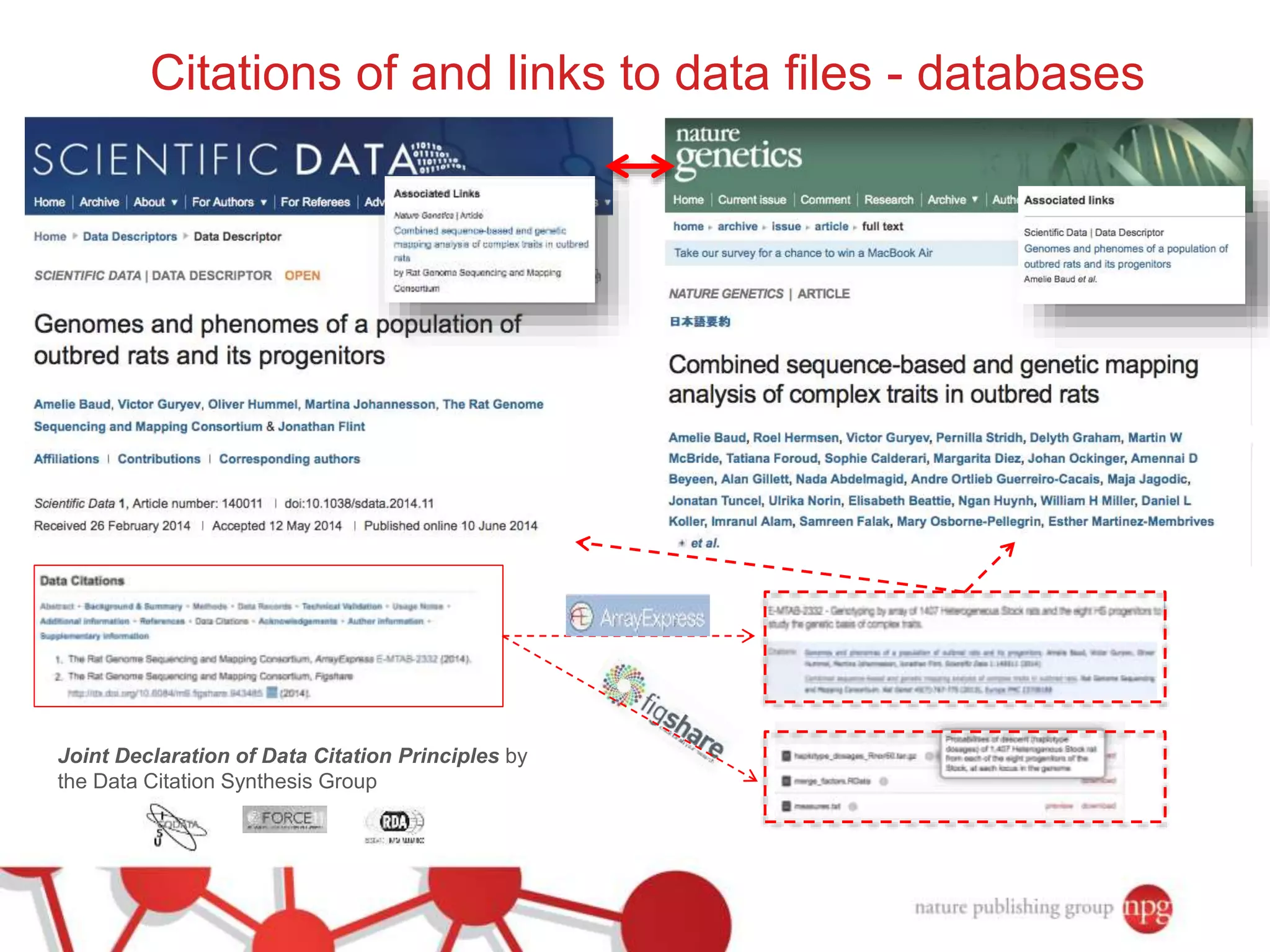
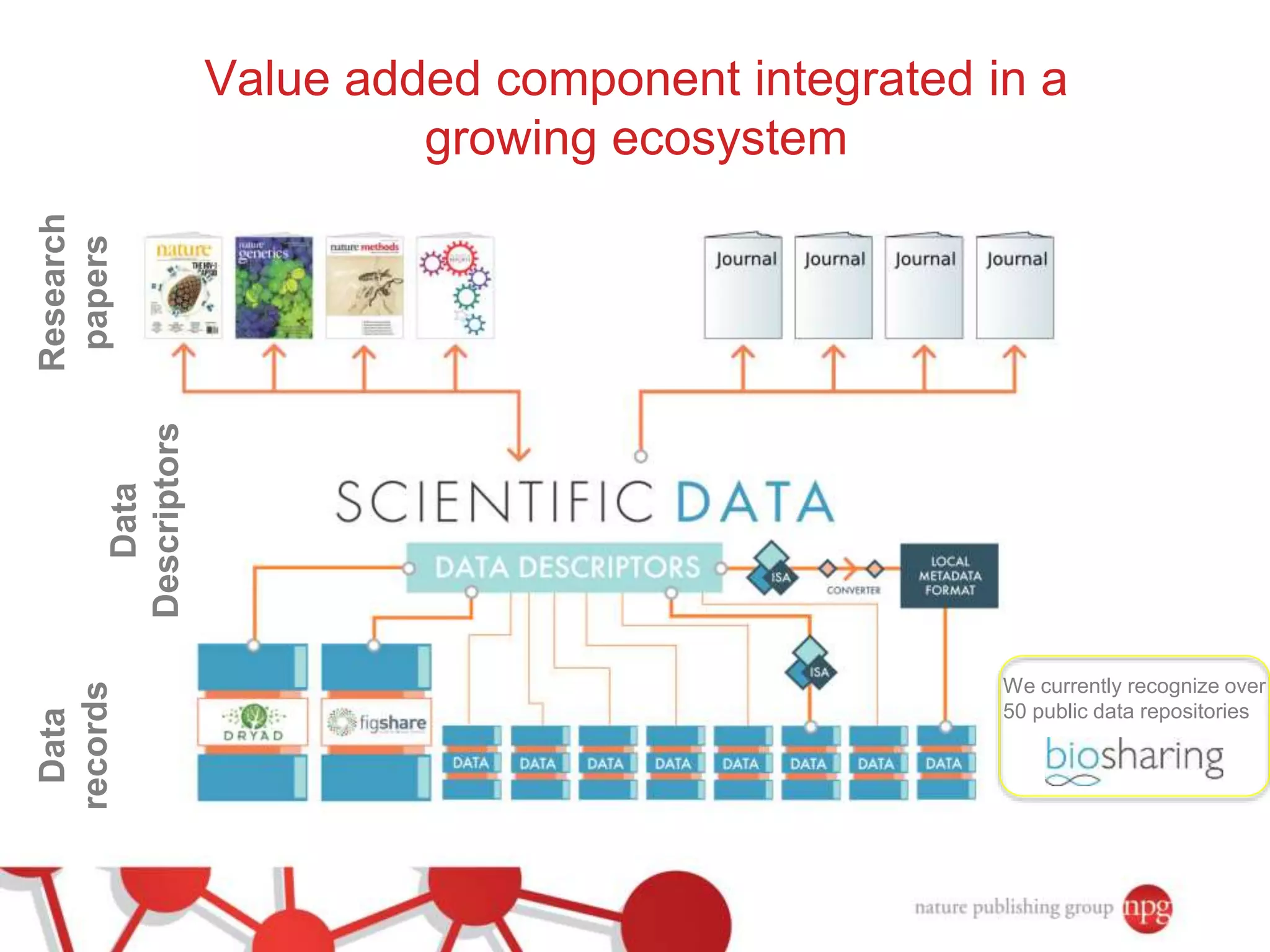
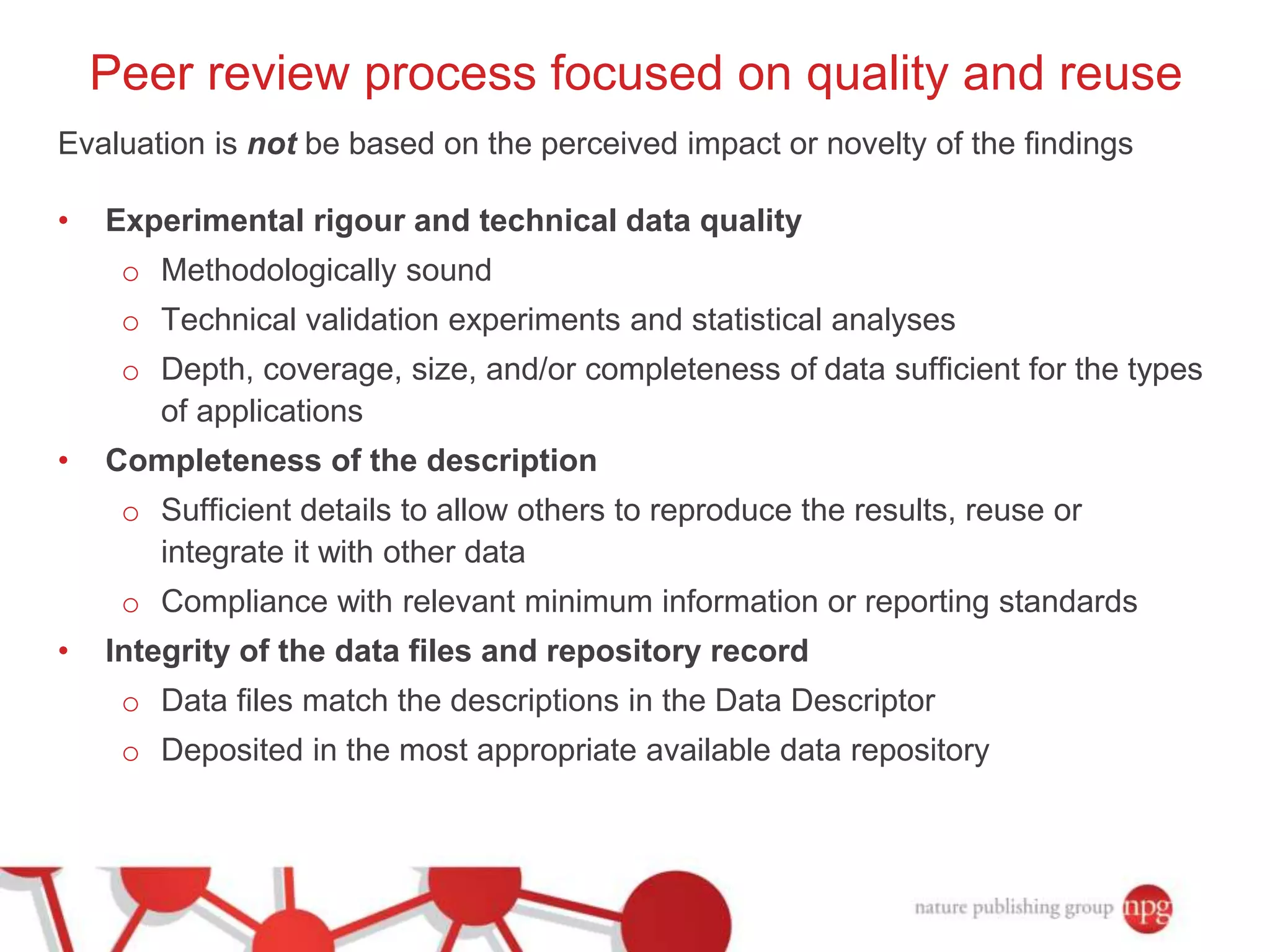
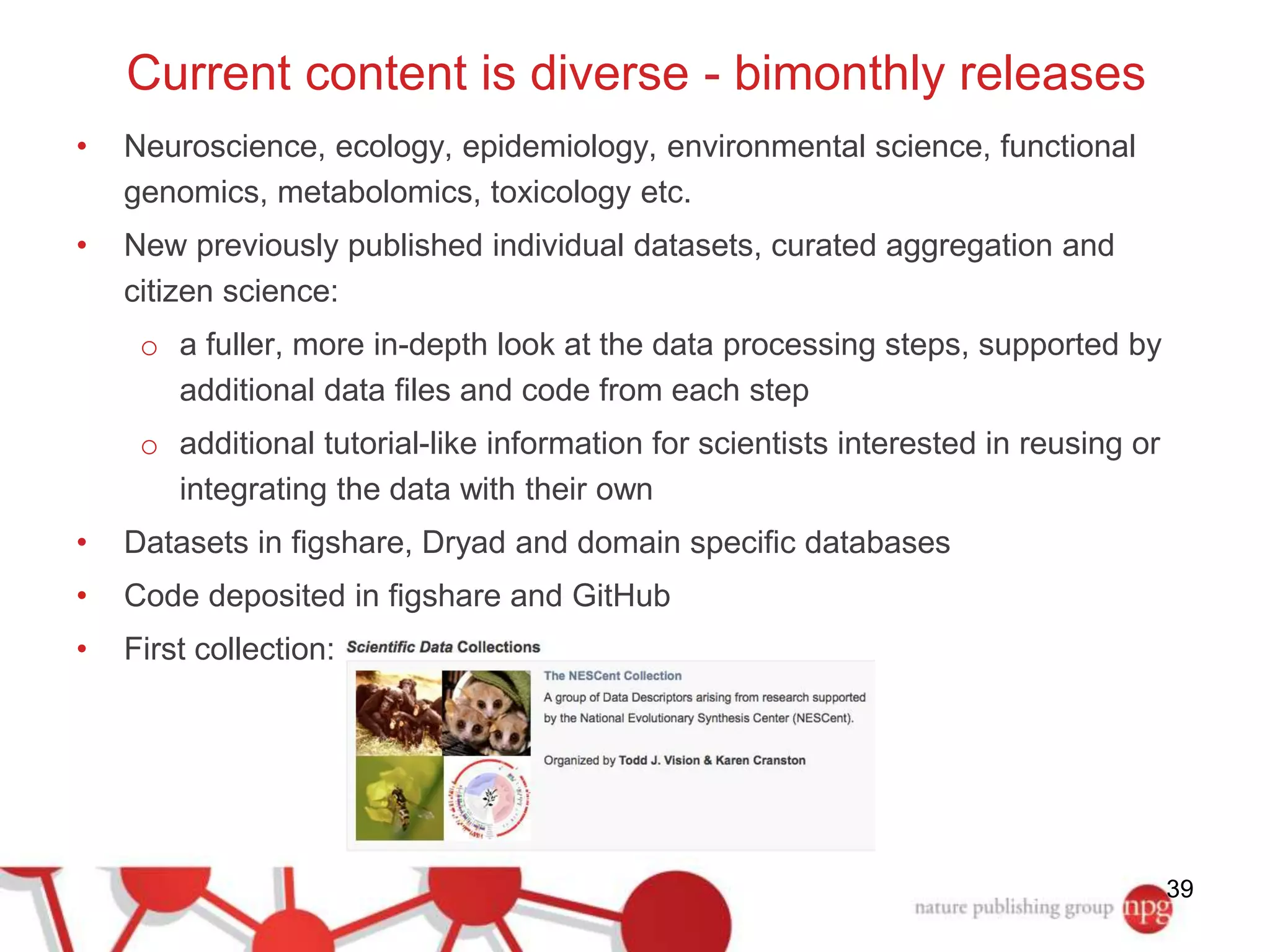
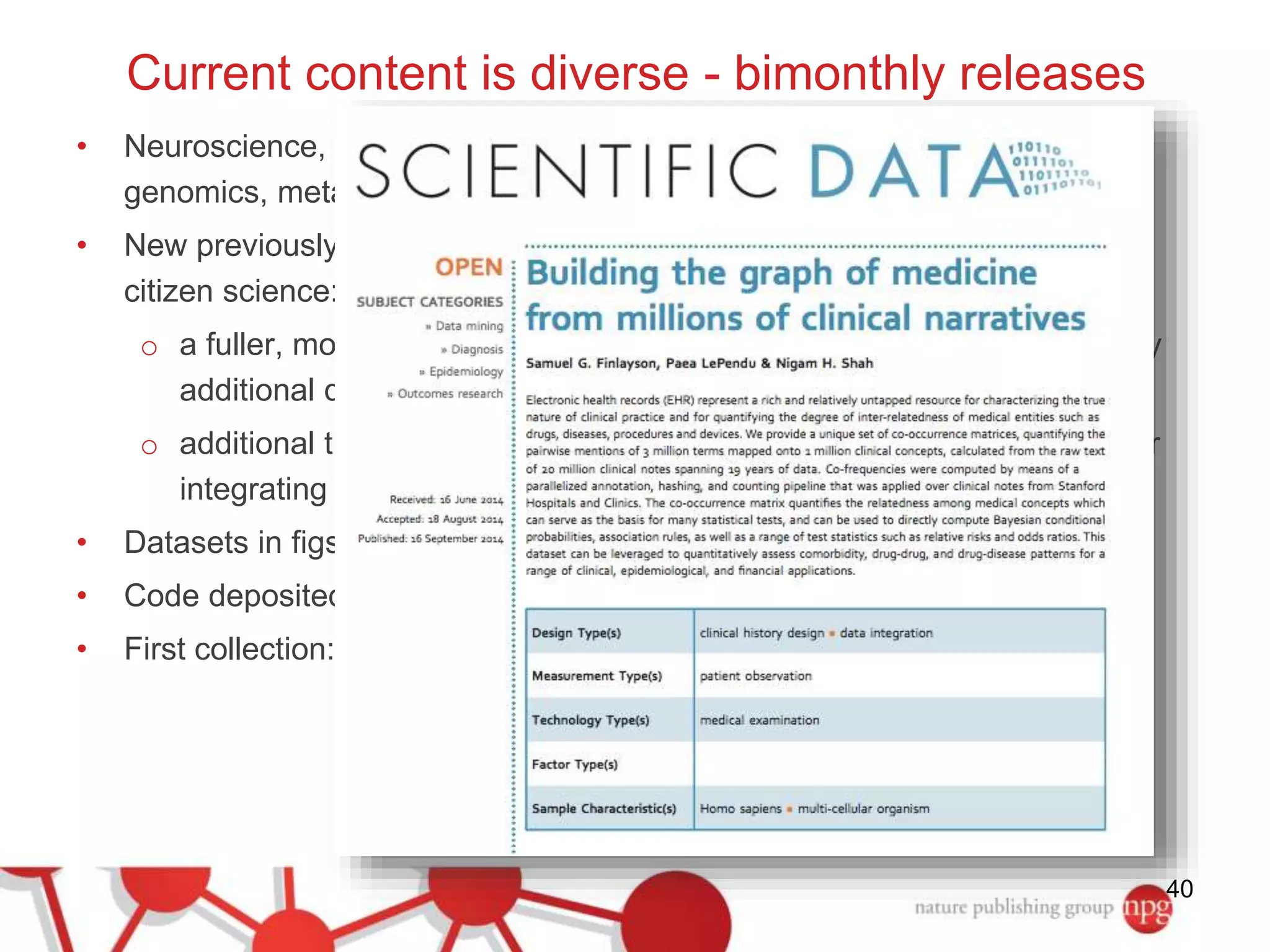
3) The document also discusses Scientific Data, a peer-reviewed journal launched by Nature Publishing Group to publish detailed descriptions of scientifically valuable datasets to facilitate reuse.