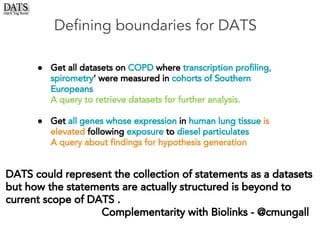
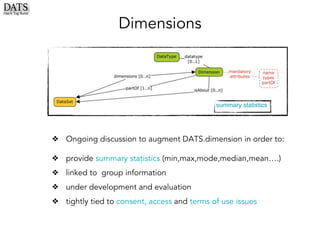
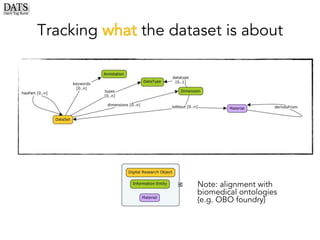
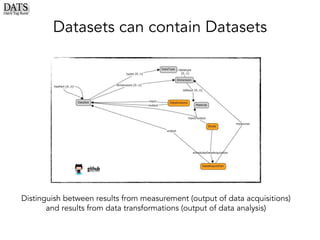
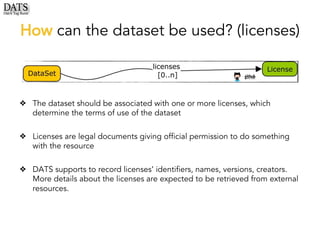
The document outlines the development of the Data Tag Suite (DATS) as part of a community initiative supported by the NIH, describing its aim to enhance the discoverability and description of datasets through a structured metadata model. It presents the iterative development process, key metadata descriptors, and the relationship between experimental details and dataset access. Additionally, the document details the integration of various metadata standards and ontologies to improve dataset indexing and accessibility for users.





![Capturing the nature of a dataset
Database of Reference Knowledge
Storing knowledge about “The building blocks”
Archive of Experiments
Storing “The signal”
[acquisition, analysis, reverse engineering]](https://image.slidesharecdn.com/dats-nih-dccpc-kc7-april2018-prs-uoxf-180420121905/85/Dats-nih-dccpc-kc7-april2018-prs-uoxf-9-320.jpg)









![Dimension: an example
{ "@type": "Dimension",
"identifier": {
"identifier": "AQ5",
"identifierSource": ""
} ,
"name": {
"valueIRI": "",
"value": "Current marital status"
},
"types":[{"value":"categorical","valueIRI":""}],
"values": [
"1 Married",
"2 Widowed",
"3 Separated",
"4 Divorced",
"5 Never married",
"-9 Missing"
],
"partOf": [
"Dataset-33581-0001.json"
],
"extraProperties": [
{
"category": "landingPage",
"values": [
"http://www.icpsr.umich.edu/
icpsrweb/ICPSR/ssvd/studies/
33581/datasets/0001/variables/AQ5"
]
}
]
}
/json-instances/ICPSR-33581/Dimension-33581-0001-AQ5.json
Credits to Matthew Richardson / Sanda Ionescu (ICPSR)](https://image.slidesharecdn.com/dats-nih-dccpc-kc7-april2018-prs-uoxf-180420121905/85/Dats-nih-dccpc-kc7-april2018-prs-uoxf-23-320.jpg)








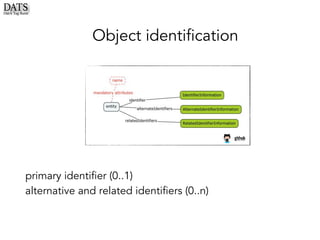

![Object identification: guidelines
master/examples/Uniprot-P77967.json
"identifier":
{
"identifier": "uniprot:P77967",
"identifierSource": "uniprot"
},
“alternateIdentifiers”:[
{
"alternateIdentifier": "PIR:S74805",
"identifierSource": "PIR"
}
],
”relatedIdentifiers": [
{
"identifier": "PANTHER:PTHR11455:SF22",
"identifierSource": “PANTHER”,
"relationType": “family and domain database” }]
❖ Primary identifier of the dataset -
can be a string, but ideally an IRI.
The identifier source is
organization/namespace
responsible for creating/hosting it
(here using “compact URIs”)
❖ Identifiers of the dataset, other
than the primary and their sources
❖ Identifiers of related resources:
useful to allow cross-references
with other complementary
resources](https://image.slidesharecdn.com/dats-nih-dccpc-kc7-april2018-prs-uoxf-180420121905/85/Dats-nih-dccpc-kc7-april2018-prs-uoxf-38-320.jpg)

![❖ Distinction between primary publication(s) and other citations
❖ If published work, pubmed or DOI will suffice
✧ Rely on dedicated APIs to recover necessary publication metadata for
indexing/search, which can be included in DATS automatically
"primaryPublications" : [
{
"identifier":
{
"identifier": "https://www.ncbi.nlm.nih.gov/pubmed/7762914",
"identifierSource": "pubmed"
},
"alternateIdentifiers": [
{
"identifier": "http://dx.doi.org/10.7326/0003-4819-123-1-199507010-00007",
"identifierSource": "doi"
}
],
}
]
Tracking bibliographic information](https://image.slidesharecdn.com/dats-nih-dccpc-kc7-april2018-prs-uoxf-180420121905/85/Dats-nih-dccpc-kc7-april2018-prs-uoxf-41-320.jpg)
![❖ Validators / Schema compliance testing
❖ DataMed Transformation Language
https://biocaddie.org/sites/default/files/d7/project/1869/biocaddie-ahm-ingestion-2017sep.pdf
[Jeff Grethe]
Tools to handle DATS documents
https://github.com/biocaddie/WG3-MetadataSpecifications](https://image.slidesharecdn.com/dats-nih-dccpc-kc7-april2018-prs-uoxf-180420121905/85/Dats-nih-dccpc-kc7-april2018-prs-uoxf-42-320.jpg)