August 2021 - JBEI Research Highlights
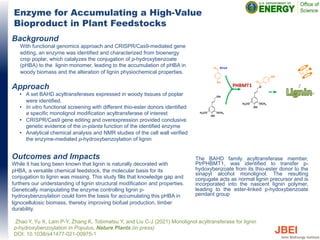
- 1. Enzyme for Accumulating a High-Value Bioproduct in Plant Feedstocks Background With functional genomics approach and CRISPR/Cas9-mediated gene editing, an enzyme was identified and characterized from bioenergy crop poplar, which catalyzes the conjugation of p-hydroxybenzoate (pHBA) to the lignin monomer, leading to the accumulation of pHBA in woody biomass and the alteration of lignin physiochemical properties. Approach • A set BAHD acyltransferases expressed in woody tissues of poplar were identified. • In vitro functional screening with different thio-ester donors identified a specific monolignol modification acyltransferase of interest • CRISPR/Cas9 gene editing and overexpression provided conclusive genetic evidence of the in-planta function of the identified enzyme • Analytical chemical analysis and NMR studies of the cell wall verified the enzyme-mediated p-hydroxybenzoylation of lignin Outcomes and Impacts While it has long been known that lignin is naturally decorated with pHBA, a versatile chemical feedstock, the molecular basis for its conjugation to lignin was missing. This study fills that knowledge gap and furthers our understanding of lignin structural modification and properties. Genetically manipulating the enzyme controlling lignin p- hydroxybenzoylation could form the basis for accumulating this pHBA in lignocellulosic biomass, thereby improving biofuel production, timber durability. Zhao Y, Yu X, Lam P-Y, Zhang K, Tobimatsu Y, and Liu C-J (2021) Monolignol acyltransferase for lignin p-hydroxybenzoylation in Populus. Nature Plants (in press) DOI: 10.1038/s41477-021-00975-1 The BAHD family acyltransferase member, PtrPHBMT1, was identified to transfer p- hydoxybenzoate from its thio-ester donor to the sinapyl alcohol monolignol. The resulting conjugate acts as normal lignin precursor and is incorporated into the nascent lignin polymer, leading to the ester-linked p-hydoxybenzoate pendant group O OCH3 OH H3CO O OH OH O SCoA OH OCH3 OH H3CO PHBMT1 O O O H
- 2. Ionic liquid-water mixtures enhance pretreatment and anaerobic digestion of agave bagasse Background • Agave bagasse (AB) is an agricultural waste generated in large quantities and an emerging alternative for production of biofuels bioproducts • Although there are several pretreatment types, using certain ionic liquids (IL) has become attractive due to its ability to dissolve the lignocellulosic biomass under mild conditions of time and temperature Approach • This work aimed to evaluate the effect of pretreatment with three diluted ILs on the A. tequilana bagasse structure and sugar yields, as well as the potential of the enzymatic hydrolysate to produce methane in batch mode • Evaluated the ILs 1-ethyl-3-methylimidazolium acetate [Emim][OAc], choline lysinate [Ch][Lys] and ethanolamine acetate [EOA][OAc] • Monitor sugar yields and develop mass-energy balances for each biomass-IL combination Outcomes and Impacts • The results showed that the IL with the best performance was [Ch][Lys], which not only offered the highest yields of sugar production (0.57 ± 0.03 g total sugars / g bagasse) but that it is possible to use it at 30% aqueous solution • Enzymatic hydrolysis of IL pretreated AB achieved 50.7 kg sugars and 49.3 kg of sugars for 90%-[Ch][Lys] and 30%-[Ch][Lys], respectively, per 100 kg of untreated biomass • The enzymatic hydrolysate from the 30 %-[Ch][Lys] pretreated AB was able to achieve 0.28 L CH4/g CODfed Perez-Pimienta et al. (2021) Industrial Crops & Products, doi: 10.1016/j.indcrop.2021.113924 Effect of agave bagasse pretreatment with IL-water mixtures on solids recovery (top), sugar production (middle) and sugar yield (bottom). Error bars represent the standard deviation.
- 3. A genomic catalog of stress response genes in anaerobic fungi for applications in bioproduction Background • Anaerobic fungi are non-model fungi capable of producing a wide array of carbohydrate active enzymes (CAZymes) • In order to develop these organisms as a biotechnology platform to produce CAZymes and other bioproducts, it is important to characterize native stress pathways such as the unfolded protein response (UPR) and heat shock response (HSR) that may be triggered by bioproduction. Approach • High-quality genomes of four strains of anaerobic fungi were mined for the components of the unfolded protein response and heat shock repsonse. • The transcriptomes of thermally stressed anaerobic fungi were sequenced and used to identify transcripts of heat shock proteins. Outcomes and Impacts • Key genes in the unfolded protein response and heat shock response were identified in the genomes of anaerobic fungi. • Anaerobic fungi mostly transcribe low molecular weight heat shock proteins (HSPs) that may help stabilize an asparagine-rich proteome. • Future engineering efforts can focus on alleviating these stress responses during bioproduction using strategies similar to those that have proven successful in model fungi such as yeasts. Swift et al. (2021) Frontiers in Fungal Biology, doi: 10.3389/ffunb.2021.708358 The genomes of anaerobic fungi encode a greater percentage of heat shock proteins compared to most other fungi, including those commonly used in bioprocessing.
- 4. Suppression of miR168 improves yield, flowering time and immunity Background • MicroRNA168 (miR168) is a key miRNA that targets Argonaute1 (AGO1), a major component of the RNA-induced silencing complex. • miR168 expression is responsive to fungal infection. • How miR168 regulates immunity to infection and whether it affects plant development was previously unknown. Approach • We found that the suppression of miR168 by a target mimic (MIM168) improves grain yield, shortens flowering time and enhances immunity to fungal infection. • These results were validated through repeated tests in rice fields in the absence and presence of the fungus. • The miR168–AGO1 module regulates miR535 to improve yield by increasing panicle number, miR164 to reduce flowering time, and miR1320 and miR164 to enhance immunity. Outcomes and Impacts • Our discovery demonstrates that changes in a single miRNA enhance the expression of multiple agronomically important traits. • Because miR168 is widely conserved in diverse plant species, including bioenergy crops, these results provide a strategy for engineering bioenergy crops for sustainable agriculture. Wang, H., Li, Y., Chern, M., Zhu Y., Zhang L.L., Lu J.H., Li X.P., Dang W.Q., Ma X.C., Yang Z.R., Yao S.Z., Zhao Z.X., Fan J., Huang Y.Y., Zhang J.W., Pu M., Wang J., He M., Li W.T., Chen X.W., Wu X.J., Li S.G., Li P., Li Y., Ronald P.C., Wang W.M. (2021) “Suppression of rice miR168 improves yield, flowering time and immunity.” Nat. Plants. A. Gross morphology and husked grains of the NPB control plant, miR168 target mimic (MIM168) transgenic lines and overexpression (OX168) lines B. Model for miR168–AGO1 regulating rice immunity and growth through three miRNAs A B
- 5. Generation of Pseudomonas putida KT2440 Strains with Efficient Utilization of Xylose and Galactose via Adaptive Laboratory Evolution Background • While P. putida KT2440 has great potential for bioprocesses, its inability to utilize the biomass abundant sugars xylose and galactose has limited its applications. Approach • Heterologous enzymes (xylD and galETKM) were introduced to construct the catabolic pathways for xylose and galactose. • Multifaceted ALE strategies were applied depending on initial growth of the engineered strains on these sugars. Outcomes and Impacts • (i) We successfully optimized poor starting strain growth (<0.1 h−1 or none) via ALE to rates of up to 0.25 h−1 on xylose and 0.52 h−1 on galactose. • (ii) Whole-genome sequencing, transcriptomic analysis, and growth screens revealed significant roles of kguT encoding a 2-ketogluconate operon repressor and 2-ketogluconate transporter, and gtsABCD encoding an ATP-binding cassette (ABC) sugar transporting system in xylose and galactose growth conditions, respectively. • (iii) We demonstrated 3.2 g/L and 2.2 g/L of indigoidine production from 10 g/L of either xylose or galactose by using the evolved strains as microbial platforms. • (iv) The generated KT2440 strains have the potential for broad application as optimized platform chassis to develop efficient microorganism-based biomass-utilizing bioprocesses. Lim et al. (2021) ACS Sustainable Chemistry & Engineering https://doi.org/10.1021/acssuschemeng.1c03765 Overall approach to enable xylose and galactose utilization DNA/RNA-seq analysis to understand mechanisms
- 6. Background • Filamentous fungi are excellent lignocellulose degraders, which they achieve through producing carbohydrate active enzymes (CAZymes). CAZyme production is highly orchestrated and gene expression analysis has greatly expanded understanding of this important biotechnological process. • Thermophilic fungus Thermoascus aurantiacus secretes highly active thermostable enzymes that enable saccharifications at higher temperatures; however, the genome-wide measurements of gene expression in response to CAZyme induction are not understood. Approach • In this study, cultivation conditions were established to perform gene expression experiments on the T. aurantiacus response to C5 and C6 sugars derived from biomass. Outcomes and Impacts • A fed-batch system with plant biomass-derived sugars D-xylose, L- arabinose and cellobiose established that these sugars induce CAZyme expression in T. aurantiacus. The C5 sugars induced both cellulases and hemicellulases, while cellobiose specifically induced cellulases (Figure 1). • It was found that d-xylose and L-arabinose strongly induced a wide variety of CAZymes, auxiliary activity (AA) enzymes and carbohydrate esterases (CEs), while cellobiose facilitated lower expression of mostly cellulase genes. • This work provides paths to produce broad spectrum thermotolerant enzymatic mixtures. Gabriel et al. (2021) Biotechnology for Biofuels, 14, 169 doi: 10.1186/s13068-021-02018-5 Figure 1. Response of T. aurantiacus to C5 and C6 sugars CAZymes from the thermophilic fungus Thermoascus aurantiacus are induced by C5 and C6 sugars
- 7. Faster, Better, and Cheaper: Harnessing Microfluidics and Mass Spectrometry for Biotechnology Background • High-throughput screening technologies are widely used for elucidating biological activities. These typically require trade-offs in assay specificity and sensitivity to achieve higher throughput. • Microfluidic approaches enable rapid manipulation of small volumes and have found a wide range of applications in biotechnology providing improved control of reaction conditions, faster assays, and reduced reagent consumption. The integration of mass spectrometry with microfluidics has the potential to create high-throughput, sensitivity, and specificity assays. Approach • This review highlights recent microfluidic approaches that have been successfully integrated with mass spectrometry analysis for high-throughput bioassays to improve throughput, sensitivity, and specificity. This review introduces the major ionization techniques of mass spectrometry that have been integrated with various microfluidic systems. • Additionally, this work discusses recent applications and shares future outlooks on multiple aspects of these technologies. Outcomes and Impacts • The major ionization techniques of mass spectrometry have been successfully integrated with various microfluidic systems for high- throughput biological assays. • Recently, high-impact applications in single-cell analysis, compound screening, and the study of microorganisms have been demonstrated. • Broad adoption of these techniques will require robust implementation of strategies for the stable sample storage, containment, and sample tracking. Ha et al. (2021) RSC Chemical Biology, DOI: 10.1039/d1cb00112d Integration of mass spectrometry with microfluidics for high-throughput bioassays (Article & image are featured on the journal back cover)
- 8. Production Cost and Carbon Footprint of Biomass-Derived Dimethylcyclooctane as a High-Performance Jet Fuel Blendstock Background • Near-term decarbonization of aviation requires energy-dense, renewable liquid fuels. • Biomass-derived 1,4-dimethylcyclooctane (DMCO), a cyclic alkane with a volumetric net heat of combustion up to 9.2% higher than Jet A, has the potential to serve as a low-carbon, high-performance jet fuel blendstock that may enable paraffinic bio-jet fuels to operate without aromatic compounds. Approach • We developed detailed process configurations for DMCO production to estimate the minimum selling price and life-cycle greenhouse gas (GHG) footprint considering three different hydrogenation catalysts and two bioconversion pathways. All modeling was done in SuperPro Designer and life-cycle GHG inventory used our previously-developed BioC2G model Outcomes and Impacts • The platinum-based catalyst offers the lowest production cost and GHG footprint of $9.0/L-Jet-Aeq and 61.4 gCO2e/MJ, given the current state of technology. • When the supply chain and process are optimized, hydrogenation with a Raney nickel catalyst is preferable, resulting in a $1.5/L-Jet-Aeq cost and 18.3 gCO2e/MJ GHG footprint if biomass sorghum is the feedstock. • Because increased gravimetric energy density of jet fuels translates to reduced aircraft weight, DMCO also has the potential to improve aircraft efficiency, particularly on long-haul flights. Baral et al. (2021) ACS Sustain. Chem. Eng., doi: 10.1021/acssuschemeng.1c03772 Figure 2. Minimum selling price of DMCO under different scenarios: (a) current state of technology (SOT) with the MVA pathway; (b) improved MVA pathway with 90% of the theoretical isoprenol yield; (c) improved MEP pathways with 90% of the theoretical isoprenol yield; and (d) optimal future case with the MVA pathway. Figure 1. Overview of 1,4-dimethylcyclooctane (DMCO) synthesis processes from the biomass-derived glucose and xylose.
- 9. A Biological Parts Search Portal and Updates to the ICE Parts Registry Software Platform Background • Modern biological part registries need additional features to address the needs of advances in synthetic biology. • The synthetic biology community will benefit from a tool providing easy accessibility to the large amounts of biological part data available in the public domain. Approach • We updated the ICE parts registry platform to include several significant new features. • We also developed a new search portal (bioparts.org) that indexes publicly available biological parts and enables searching using keywords or sequence fragments (Figure 1). Outcomes and Impacts • We highlighted the significant features that were developed for ICE since its introduction over a decade ago. This includes the implementation of the web of registries concept which creates a single distributed parts database out of multiple independent ICE instances. • We established a search portal that sources data from registries and repositories such as IGEM, NCBI GenBank, SynbioHub, ICE, AddGene and Genscript (Figure 2). The availability of an API enables third-party applications to utilize features such as automatic sequence annotation. Plahar, et al., “BioParts - A Biological Parts Search Portal and Updates to the ICE Parts Registry Software Platform”, ACS Synthetic Biology. doi: 10.1021/acssynbio.1c00263 Figure 1 Figure 2
- 11. Plant single-cell solutions for energy and the environment Background • Single-cell transcriptomics has led to fundamental new insights into animal biology, such as the discovery of new cell types and cell type-specific disease processes • The application of single-cell approaches to plants, fungi, algae, or bacteria (environmental organisms) has been far more limited, largely due to the challenges posed by polysaccharide walls surrounding these species’ cells Approach • In this perspective, we discuss opportunities afforded by single-cell technologies for energy and environmental science and grand challenges that must be tackled to apply these approaches to plants, fungi and algae • Highlight the need to develop better and more comprehensive single-cell technologies, analysis and visualization tools, and tissue preparation methods Outcomes and Impacts • advocate for the creation of a centralized, open-access database to house plant single-cell data. • These efforts should balance the need for deep characterization of select model species while still capturing the diversity in the plant kingdom. Investments into the development of methods, their application to relevant species, and the creation of resources to support data dissemination will enable groundbreaking insights to propel energy and environmental science forward Cole et al. (2021) Commun Biol. doi: 10.1038/s42003-021-02477-4 Using single-cell methods in bioproducts and biomaterials applications.
- 12. Co-cultivation of anaerobic fungi with rumen bacteria establishes an antagonistic relationship Background • The rumen microbiome efficiently degrades lignocellulosic biomass through its wide array of bacterial and fungal carbohydrate active enzymes (CAZymes). • Little is known about how these microorganisms cooperate or compete in order to degrade biomass. • Fungal interactions with rumen bacteria are not well characterized. Approach • Synthetic co-cultures of anaerobic fungi and rumen bacteria were cultivated on switchgrass (a bioenergy crop) and crystalline cellulose (Avicel® ) for comparison. • This study uses ‘omics’ technologies (genomics, transcriptomics, and metabolomics) to illustrate the antagonistic relationship between anaerobic fungi and bacteria. Outcomes and Impacts • Biosynthetic genes of fungal secondary metabolites were upregulated during bacterial challenge, and rumen bacteria responded by upregulating drug efflux pumps and toxin-antitoxin systems. • Transcriptional studies and western blotting affirmed that the secondary metabolism of anaerobic fungi is likely regulated by chromatin remodeling. • The fundamental understanding provided by this study is an important step toward engineering synthetic microbial consortia for biomass degradation. Swift et al. (2021) mBio, doi: 10.1128/mBio.01442-21 Helium ion microscopy of the anaerobic fungus Anaeromyces robustus and the bacteria Fibrobacter UWB7 illustrates close contact of these rumen microorganisms in a synthetic co- culture.
- 13. Plant genome engineering from lab to field— a Keystone Symposia report Background • Enhancing crop resiliency and yield via genome engineering will be a key part of tackling food source challenges posed by a growing global population and a warming climate. • New tools such as CRISPR/Cas have ushered in significant advances in plant genome engineering, but several challenges remain, including efficient transformation and plant regeneration for most crop species, low frequency of some editing applications, and high attrition rates. Approach • Experts in plant genome engineering and breeding from academia and industry met virtually for the Keystone eSymposium “Plant Genome Engineering: From Lab to Field” to discuss advances in genome editing tools, plant transformation, plant breeding, and crop trait development, all vital for transferring the benefits of novel technologies to the field. Outcomes and Impacts • Presenters described novel tools for gene and base editing, gene targeting by homology-directed repair, transcriptional regulation, epigenetic editing, and induced chromosomal rearrangements. • Presenters also discussed applications of genome editing in agriculture, including improving potato traits with CRISPR/Cas, improving cereal crop transformation, and increasing fruit and vegetable consumption with genome editing. Cable et al. (2021) Annals of the New York Academy of Sciences, doi: 10.1111/nyas.14675 A comprehensive analysis of plant core promoters.