JBEI June 2019 highlight slides
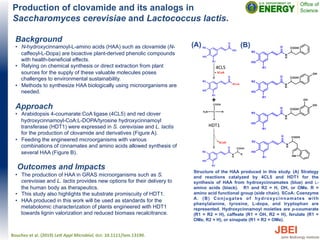
- 1. Production of clovamide and its analogs in Saccharomyces cerevisiae and Lactococcus lactis. Background • N-hydroxycinnamoyl-L-amino acids (HAA) such as clovamide (N- caffeoyl-L-Dopa) are bioactive plant-derived phenolic compounds with health-beneficial effects. • Relying on chemical synthesis or direct extraction from plant sources for the supply of these valuable molecules poses challenges to environmental sustainability. • Methods to synthesize HAA biologically using microorganisms are needed. Approach • Arabidopsis 4-coumarate:CoA ligase (4CL5) and red clover hydroxycinnamoyl-CoA:L-DOPA/tyrosine hydroxycinnamoyl transferase (HDT1) were expressed in S. cerevisiae and L. lactis for the production of clovamide and derivatives (Figure A). • Feeding the engineered microorganisms with various combinations of cinnamates and amino acids allowed synthesis of several HAA (Figure B). Outcomes and Impacts • The production of HAA in GRAS microorganisms such as S. cerevisiae and L. lactis provides new options for their delivery to the human body as therapeutics. • This study also highlights the substrate promiscuity of HDT1. • HAA produced in this work will be used as standards for the metabolomic characterization of plants engineered with HDT1 towards lignin valorization and reduced biomass recalcitrance. Bouchez et al. (2019) Le$ Appl Microbiol, doi: 10.1111/lam.13190. Structure of the HAA produced in this study. (A) Strategy and reactions catalyzed by 4CL5 and HDT1 for the synthesis of HAA from hydroxycinnamates (blue) and L- amino acids (black). R1 and R2 = H, OH, or OMe. R = amino acid functional group (side chain). SCoA: Coenzyme A. (B) Conjugates of hydroxycinnamates with phenylalanine, tyrosine, L-dopa, and tryptophan are represented. Hydroxycinnamoyl moieties are p-coumarate (R1 = R2 = H), caffeate (R1 = OH, R2 = H), ferulate (R1 = OMe, R2 = H), or sinapate (R1 = R2 = OMe). (A) (B)
- 2. Sphingolipid biosynthesis modulates plasmodesmal ultrastructure and phloem unloading Background • Phloem loading/unloading is critical for the transport of the products of photosynthesis from the source tissue (e.g. leaves) to sink tissue (e.g. roots). • Plasmodesmata are channels which cross the plant cell wall and allow cell-to cell transport and communication. These are lined with plasma membrane, and contain the desmotubule – a narrow tube that connects the smooth endoplasmic reticulum of the two cells. They come in two forms (Type I and Type II). • The cytoplasm between the desmotubule and the plasma membrane allows for symplastic transport. It is not clear how this is regulated. Approach • In work led by researchers at the Universities of Cambridge (UK) and Bordeaux (France), a mutant in sphingolipid biosynthesis (PHLOEM UNLOADING MODULATOR, PLM) was identified via a suppressor screen of a callose gain-of-function mutant (cals3-d) with aberrant plasmodesmata function. Outcomes and Impacts • plm had decreased GIPCs and ceramides (sphingolipids) in the plasma membrane • PLM has a role in regulating plasmodesmata membrane composition, and leads to a change in plasmodesmata type. • In turn, this alters plasmodesmata permeability. • This demonstrates that plasmodesmata are not regulated by the cell wall (or callose) alone. Yan et al. (2019) Nature Plants, 5:604-‐615, doi: 10.1038/s41477-‐019-‐0429-‐5 pSUC2::GFP expressed in Arabidopsis roots, which is used as a measure of phloem unloading. GIPC composiRon in wild type (Col-‐0) and plm-‐1
- 3. A web-based tool for the prediction of rice transcription factor function Background • Only a small number of genes encoding transcription factors (TFs) have been characterized in Oryza sativa (rice). • Ancient genome duplications indicate that ~50% of all genes related to non-transposable elements in rice are functionally redundant. • Gene duplication and functional redundancy complicate the analysis of TFs. Approach • Focused on rice transcription factors (TFs) and transcriptional regulators (TRs). • Developed a web-based tool called the Rice Transcription Factor Phylogenomics Database (RTFDB) and demonstrated its application for predicting TF function and to identify tissue-specific and stress-related gene expression. Outcomes and Impacts • 273 genes preferentially expressed in specific tissues or organs, 455 genes showing a differential expression pattern in response to 4 abiotic stresses, 179 genes responsive to infection of various pathogens and 512 genes showing differential accumulation in response to various hormone treatments were identified. • Estimated a predominant role for 83.3% (65/78) of the TF or transcriptional regulator genes that had been characterized via loss-of-function studies. • Method is applicable for functional studies of other bioenergy crop species with annotated genomes. Chandran et al. (2019) Database, doi: 10.1093/database/baz061 Heatmaps of featured gene expression groups that are derived from meta-‐expression analysis of (A) anatomical samples, (B) abioRc stress samples, (C) bioRc stress samples and (D) hormone-‐treated samples. The number of genes per group is shown in parentheses aXer the group name. For stress and hormone-‐treated featured groups, a staRsRcal cut-‐off of >2 log2 fold-‐change at P < 0.05 was used.
- 4. Integration of renewable deep eutectic solvents with engineered biomass to achieve a closed-loop biorefinery Background • Deep eutectic solvents (DESs) have gained increasing attention due to their properties including universal solvating capabilities and wide tunability. • Ease of synthesis and broad availability from inexpensive renewable source such as lignin could render DESs more versatile solvents for biomass pretreatment. • Certain lignin mutants accumulate unusual aromatics that can be potentially converted into valuable DESs for a closed-loop biorefinery. Approach • We developed a process that integrates the use of low-recalcitrant engineered biomass with a pretreatment consisting of lignin- derived DESs. • Arabidopsis engineered plants deficient for cinnamyl alcohol dehydrogenase (CAD) accumulate lignin enriched in cinnamaldehydes instead of cinnamyl alcohols. • Renewable DESs can be synthesized from such phenolic aldehydes. Outcomes and Impacts • New DESs derived from vanillin and 4-hydroxybenzaldehyde were developed (ChCl-VAN and ChCl-HBA). • ChCl-VAN and ChCl-HBA are efficient for the pretreatment of biomass prior enzymatic saccharification. • In addition to exhibit reduced biomass recalcitrance, CAD mutants represent an ideal source of vanillin and 4-hydroxybenzaldehyde for DES synthesis. Kim et al. (2019) Proc. Natl. Acad. Sci. U.S.A., doi: 10.1073/pnas.1904636116. Complex formation in two new DESs ChCl-HBA and ChCl-VAN synthesized for biomass pretreatment. Glucan conversion from CAD mutant and wild-type (WT) biomass pretreated with the new DESs, ChCl-VAN and ChCl- HBA. R = H, hydroxybenzaldehyde (HBA) R = OCH3, vanillin (VAN)
- 5. A dominant negative approach to reduce xylan in plants Background • After cellulose, the hemicellulose xylan make up the majority of the carbohydrate content of plant biomass. • Xylan is composed of pentoses, which are difficult to convert into biofuels and bioproducts, and reduced xylan is a desirable trait of bioenergy crops. • Synthesis of the xylan backbone requires the proteins IRX9, IRX14 and IRX10 and their partially redundant homologs. Mutants in the corresponding genes have reduced xylan but compromised growth properties. Approach • Conserved residues of IRX10 xylan synthase were mutated in order to generate non-functional versions of IRX10. • Plants were transformed with the mutated IRX10 proteins under 35S promoter, with the hypothesis that a non-functional enzyme would serve as a dominant negative. Outcomes and Impacts • Mutations in glycine-238 or glutamate-293 caused strong reduction in growth. • The plants had up to 55% reduction in cell wall xylose, and strongly reduced xylan content, confirming the hypothesis • The results confirm that IRX10 functions in a protein complex • The approach can be used to generate healthy plants with decreased xylan by restricting expression of the mutated IRX10 to fiber cells • A similar dominant negative approach can be employed as a biosystems design tool in other cases where a protein complex is required for a biosynthetic reaction Brandon et al. (2019) Plant Biotechnol. Journal, doi: 10.1111/pbi.13198 Some of the plants expressing mutated IRX10 proteins have reduced growth. The residue G283 is conserved in human EXT1 protein and mutaRons are known to cause disease. It may be a catalyRc residue but the structure of IRX10 or EXT1 is not known. Xylan synthase acRvity is reduced in plants with mutated IRX10. AcRvity was determined in microsomes isolated from stems and using xylohexaose-‐ANTS as substrate. Products were analyzed by PACE. Significant differences from EVC are indicated (p < 0.01, ANOVA and Dunnef’s test). EVC IRX10 H146D F277A C278A G283D E293Q 0 100 200 300 400 500 600 700 800 900 Rha Ara Gal Glc Xyl Man GalA GlcA nmol/mgAIR Col-0 P-value IRX10 .321 H146D .012 C278A .627 G283D .002 E293Q .001 EVC 0 25 50 75 100 XylTactivity (%ofEVC) EVC IRX10 G283D E293Q Cell walls (AIR) from mutated plants have less xylose compared to empty vector control (EVC). Bars show average ± SD (n=3-‐5). The p-‐values indicate significance of difference from EVC (t-‐test). ** **
- 6. Toolset of constitutive promoters for metabolic engineering of Rhodosporidium toruloides Integration at CAR2 locus (A) Strain & Constructs Selection of white colonies EGFP mRuby CellCountCellCount (B) Transformation (C) Growth (D) Analysis R. toruloides IFO0880 ∆ku70 Promoter EGFP mRuby2 EGFPPromoter 58 constructs SD 1% glucose SD 1% xylose SD 1% xylose 1% glucose Growth in 24-well plates YPD mRuby21. 2. mRuby EGFPPROMOTER CAR2 CAR2CAR2 Bidirec'onal Background • Rhodosporidium toruloides is a promising host for the production of biofuels and bioproducts from lignocellulosic biomass. • there is a lack of characterized promoters to drive expression of heterologous genes for strain engineering in R. toruloides. Approach • Candidates selected based on RNAseq data. • 58 dual reporter constructs synthesized at the JGI. • Site-specific integration into Rhodosporidium toruloides. • Promoter expression strength was determined by measurement of EGFP and mRuby2 reporters by flow cytometry. Outcomes and Impacts • Established A set of robust and constitutive promoters to facilitate genetic engineering of R. toruloides is presented here, ranging from a promoter previously used for this purpose (P7, glyceraldehyde 3-phosphate dehydrogenase, GAPDH) to stronger monodirectional (e.g., P15, mitochondrial adenine nucleotide translocator, ANT) and bidirectional (e.g., P9 and P9R, histones H3 and H4, respectively) promoters. • This set of characterized promoters significantly expands the range of engineering tools available for this yeast and can be applied in future metabolic engineering studies for the production of biofuels and bioproducts. Nora et al. (2019) Microbial Cell Factories, doi: 10.1186/s12934-‐019-‐1167-‐0
- 7. Early cyanobacteria and the innovation of microbial sunscreens Background • Understanding when cyanobacteria evolved the ability to produce small molecules that function as sunscreens provides and understanding of when basic photoprotection mechanisms evolved. • UV photoprotection is a common adaptation across microbes and plants, with different compounds that are able to function as sunscreens. Shih et al. (2019) mBIO, doi: 10.1128/MBIO.01262-‐19 Approach • We provide a review and commentary piece on the field of microbial evolution and secondary metabolism of the cyanobacterial sunscreen, scytonemin. Outcomes and Impacts • Understanding when scytonemin biosynthesis originated provides a constraint key insight on the origins of oxygenic photosynthesis in cyanboacteria and how early phototrophic organisms had to evolve to both oxygen and light.
- 8. Xyloglucan endotransglucosylase-hydrolase 30 negatively affects salt tolerance in Arabidopsis Background • Plants have evolved various strategies to sense and respond to saline environments that severely reduce plant growth and limit agricultural productivity. Alteration to the cell wall is one strategy that helps plants adapt to salt stress. However, how cell wall components respond to salt stress is not fully understood. Approach • Performed salt sensitivity assay of xth30 mutant plants. • A series of analysis was performed, including H2O2 level measurement, antioxidant enzyme activity measurement, subcellular localization of XTH30, cellulose content, structural analysis of XyG oligosaccharides and microtubule observation. Outcomes and Impacts • XTH30, encoding a xyloglucan endotransglucosylase- hydrolase, negatively affected salt tolerance. • We revealed that XTH30 negatively affected salt tolerance, probably through modulating XyG side chains composition, affecting XLFG accumulation, cellulose synthesis and cortical microtubule stability. • The study showed that down regulation of XTH30 can be a potential strategy to increase plant drought resistance. Yan et al. (2019) Journal of Experimental Botany, doi: 10.1093/jxb/erz311 XTH30 negatively affects salt stress tolerance A potential model for XTH30 regulation in salt stress responses
- 9. A screening method to identify efficient sgRNAs in Arabidopsis, used in conjunction with cell- specific lignin reduction Background • Single guide RNA (sgRNA) selection is important for the efficiency of CRISPR/Cas9 mediated genome editing. However, in plants, the rules governing selection are not well established. Approach • Developing an assay to test sgRNA efficiency in vivo. • In case study I, use the selected sgRNA from the assay to knockout HCT, an essential gene involved in lignin biosynthesis, in a tissue-specific manner • In case study II, use the selected sgRNA from the assay to generate mutant of GONST2, a nucleotide sugar transporter involved in GIPC synthesis Outcomes and Impacts • A transient assay was developed, which is widely applicable to evaluate sgRNA efficiency before applying CRISPR genome editing in stable transgenic plants. • Using highly efficient sgRNA, chimeric hct plants were generated for the first time. The plants showed decreased lignin content and increased saccharification rate, while maintaining plant integrity. • The study also indicated that the use of highly efficient sgRNA may accelerate the process of expanding germplasm for both molecular breeding and research. Liang et al. (2019) Biotechnology for Biofuels, 12: 130, doi: 10.1186/s13068-‐019-‐1467-‐y Development of a transient assay to test sgRNA efficiency for CRISPR ediRng 20-‐30% reducRon in total lignin content (A) and 30-‐50% increase in saccharificaRon rate (B) were observed in chimeric hct plants A B