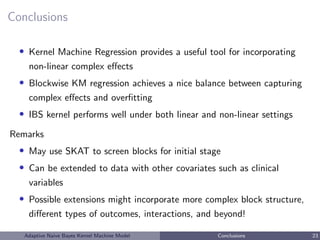
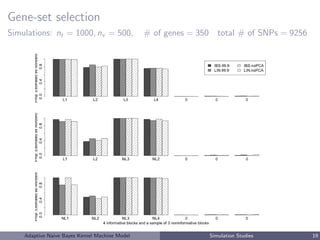
This document describes a risk classification method using an adaptive naive Bayes kernel machine model. The method partitions genetic data into blocks, such as gene sets or linkage disequilibrium blocks, and applies kernel machine regression within each block to allow for complex, nonlinear effects. Regularized selection of informative blocks is used to build an accurate yet parsimonious prediction model. Simulation studies show the method achieves high prediction accuracy and correctly selects predictive blocks. The method is applied to genetic risk prediction of type 1 diabetes using single nucleotide polymorphism data from known risk loci.







![Approach
Challenge:
• Prediction models based on univariate testing, additive models, global
methods → low prediction accuracy, low AUC, missing heritability
• Non-linear effects? testing for interactions → low power
Our approach [Minnier et al., 2015]:
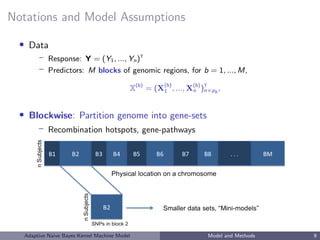
• Blockwise method:
leverage biological knowledge to build models at the gene-set level
genes, gene-pathways, linkage disequilibrium blocks
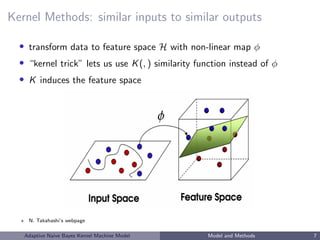
• Kernel machine regression:
allow for complex and nonlinear effects
implicitly specify underlying complex functional form of covariate
effects via similarity measures (kernels) that define the distance
between two sets of covariates
Adaptive Naive Bayes Kernel Machine Model Model and Methods 6](https://image.slidesharecdn.com/kmpresminnierjune2015-161209012516/85/Risk-Classification-with-an-Adaptive-Naive-Bayes-Kernel-Machine-Model-9-320.jpg)

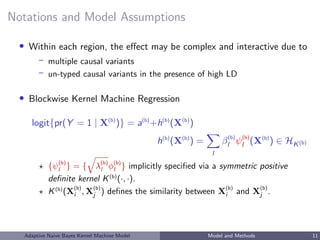
![Notations and Model Assumptions
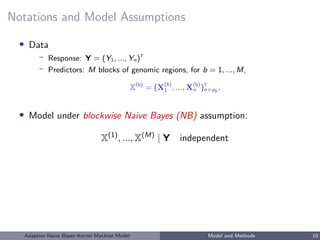
• Data
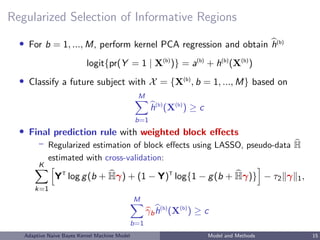
– Response: Y = (Y1, ..., Yn)T
– Predictors: M blocks of genomic regions, for b = 1, ..., M,
X(b)
= (X
(b)
1 , ..., X(b)
n )T
n×pb
,
• Model under blockwise Naive Bayes (NB) assumption:
X(1)
, ..., X(M)
| Y independent ⇒
logit{pr(Y = 1 | X(1)
, ..., X(M)
)} = c +
M
b=1
logit{pr(Y = 1 | X(b)
)}
– NB assumption allows separate estimation by block and reduces overfitting
– Performs well for zero-one loss L(X) = I( ˆY (X) = Y ) [Domingos and
Pazzani, 1997]
Adaptive Naive Bayes Kernel Machine Model Model and Methods 10](https://image.slidesharecdn.com/kmpresminnierjune2015-161209012516/85/Risk-Classification-with-an-Adaptive-Naive-Bayes-Kernel-Machine-Model-16-320.jpg)
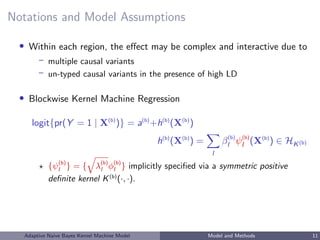
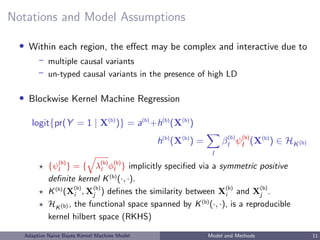
![Choices of Kernel Functions
Linear kernel: K(Xi , Xj ) = ρ + XT
i Xj ,
h(X) =
p
k=1
βkXk
Fitting logistic regression with linear kernel ⇔ logistic ridge regression.
IBS kernel: K(Xi , Xj ) = p
k=1(2 − |Xik − Xjk|),
powerful in detecting non-linear effects with SNP data [Wu et al, 2010]
Adaptive Naive Bayes Kernel Machine Model Model and Methods 12](https://image.slidesharecdn.com/kmpresminnierjune2015-161209012516/85/Risk-Classification-with-an-Adaptive-Naive-Bayes-Kernel-Machine-Model-22-320.jpg)
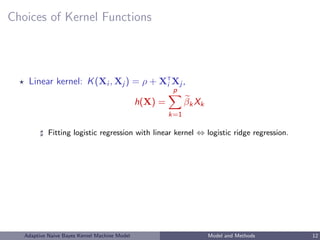
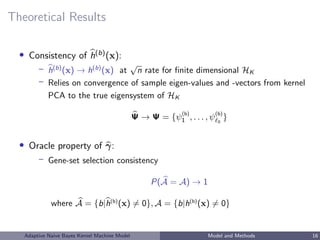
![Estimation of h: Kernel PCA
• primal form: h = l βl ψl = l βl
√
λl φl
• Kernel PCA approximation:
K = [K(Xi , Xj )]1≤i,j≤n =
n
l=1
λl φl φ
T
l
K =
0
l=1
λl φl φ
T
l = ΨΨ
T
; Ψ = [λ
1
2
1 φ1, ..., λ
1
2
0
φ 0
]n× 0
Scholkopf et al. [1999]; Williams and Seeger [2000]; Braun et al. [2008]; Zhang et al. [2010]
• h(b)
(X(b)
) = Ψβ
• obtain (a, β) as the minimizer of ridge logistic objective function
L(Y , a, Ψβ) + τ β 2
Adaptive Naive Bayes Kernel Machine Model Model and Methods 13](https://image.slidesharecdn.com/kmpresminnierjune2015-161209012516/85/Risk-Classification-with-an-Adaptive-Naive-Bayes-Kernel-Machine-Model-23-320.jpg)
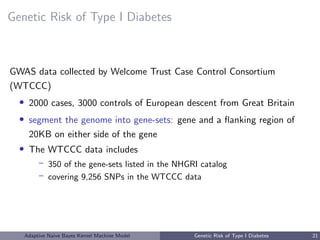
![Genetic Risk of Type I Diabetes
• Autoimmune disease, usually diagnosed in childhood
• T1D
– 75 SNPs have been identified as T1D risk alleles (National Human
Genome Research Institute, Hindorff et al. [2009])
– 91 genes that either contain these SNPs or flank the SNP on either
side on the chromosome
Adaptive Naive Bayes Kernel Machine Model Genetic Risk of Type I Diabetes 20](https://image.slidesharecdn.com/kmpresminnierjune2015-161209012516/85/Risk-Classification-with-an-Adaptive-Naive-Bayes-Kernel-Machine-Model-30-320.jpg)
![Genetic Risk of Type I Diabetes
• Autoimmune disease, usually diagnosed in childhood
• T1D
– 75 SNPs have been identified as T1D risk alleles (National Human
Genome Research Institute, Hindorff et al. [2009])
– 91 genes that either contain these SNPs or flank the SNP on either
side on the chromosome
• T1D + Other autoimmune diseases (Rheumatoid arthritis, Celiac
disease, Crohns disease, Lupus, Inflammatory bowel disease)
– 365 SNPs have been identified as T1D+other autoimmune disease risk
alleles (NHGRI)
– 375 genes that either contain these SNPs or flank the SNP on either
side on the chromosome
Adaptive Naive Bayes Kernel Machine Model Genetic Risk of Type I Diabetes 20](https://image.slidesharecdn.com/kmpresminnierjune2015-161209012516/85/Risk-Classification-with-an-Adaptive-Naive-Bayes-Kernel-Machine-Model-31-320.jpg)