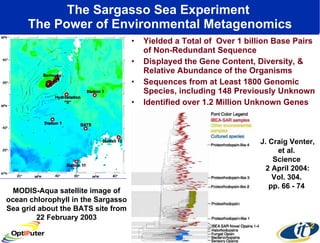
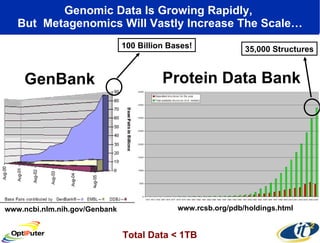
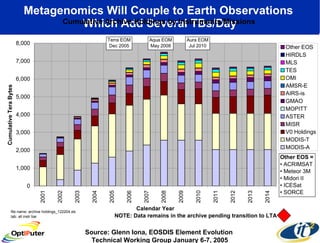
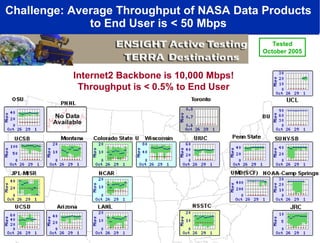
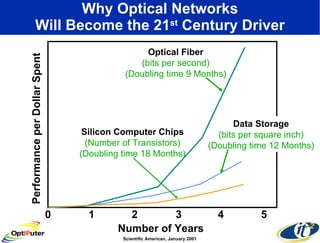
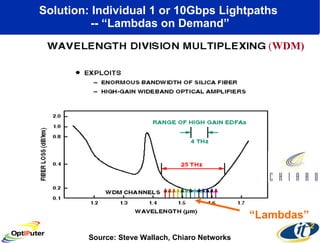
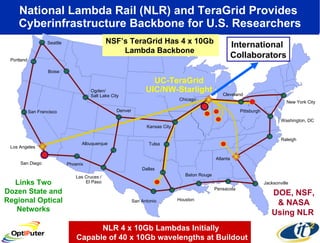
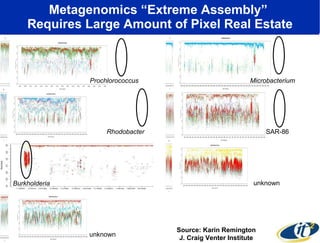
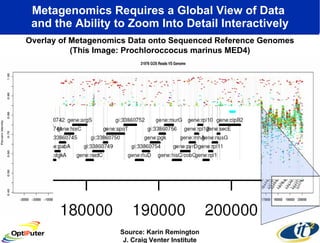
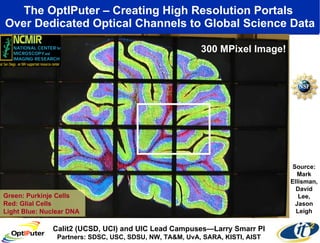
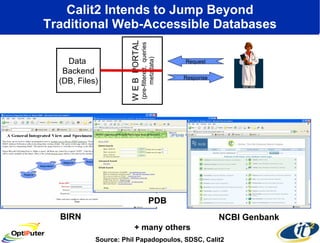
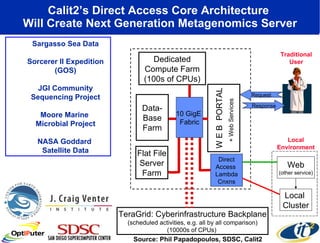
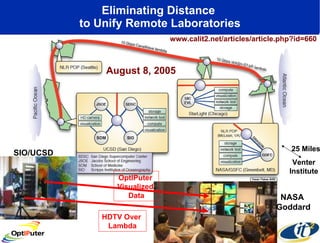
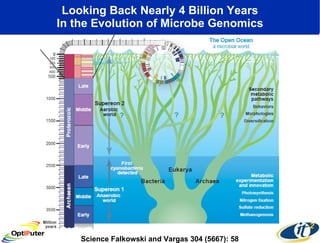
Dr. Larry Smarr's talk at UCSD highlighted advancements in metagenomics and the rapid growth of genomic data, emphasizing the need for robust information infrastructure to support this field. The Sargasso Sea Experiment demonstrated the potential of environmental metagenomics, revealing genetic diversity and discovering numerous unknown genes. Collaboration through initiatives like the National Lambda Rail and teragrid aims to enhance data accessibility and analysis for researchers across various scientific disciplines.