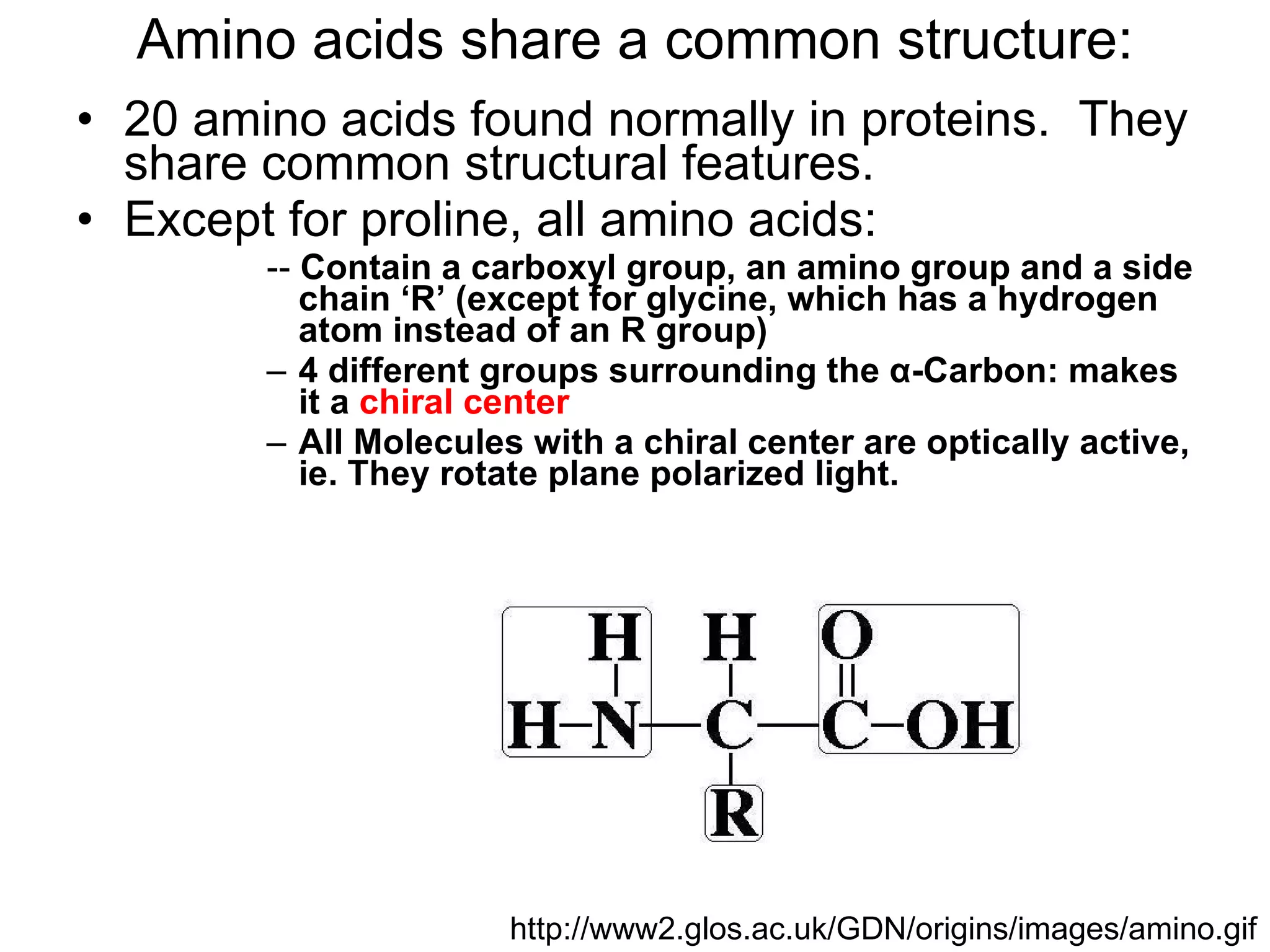
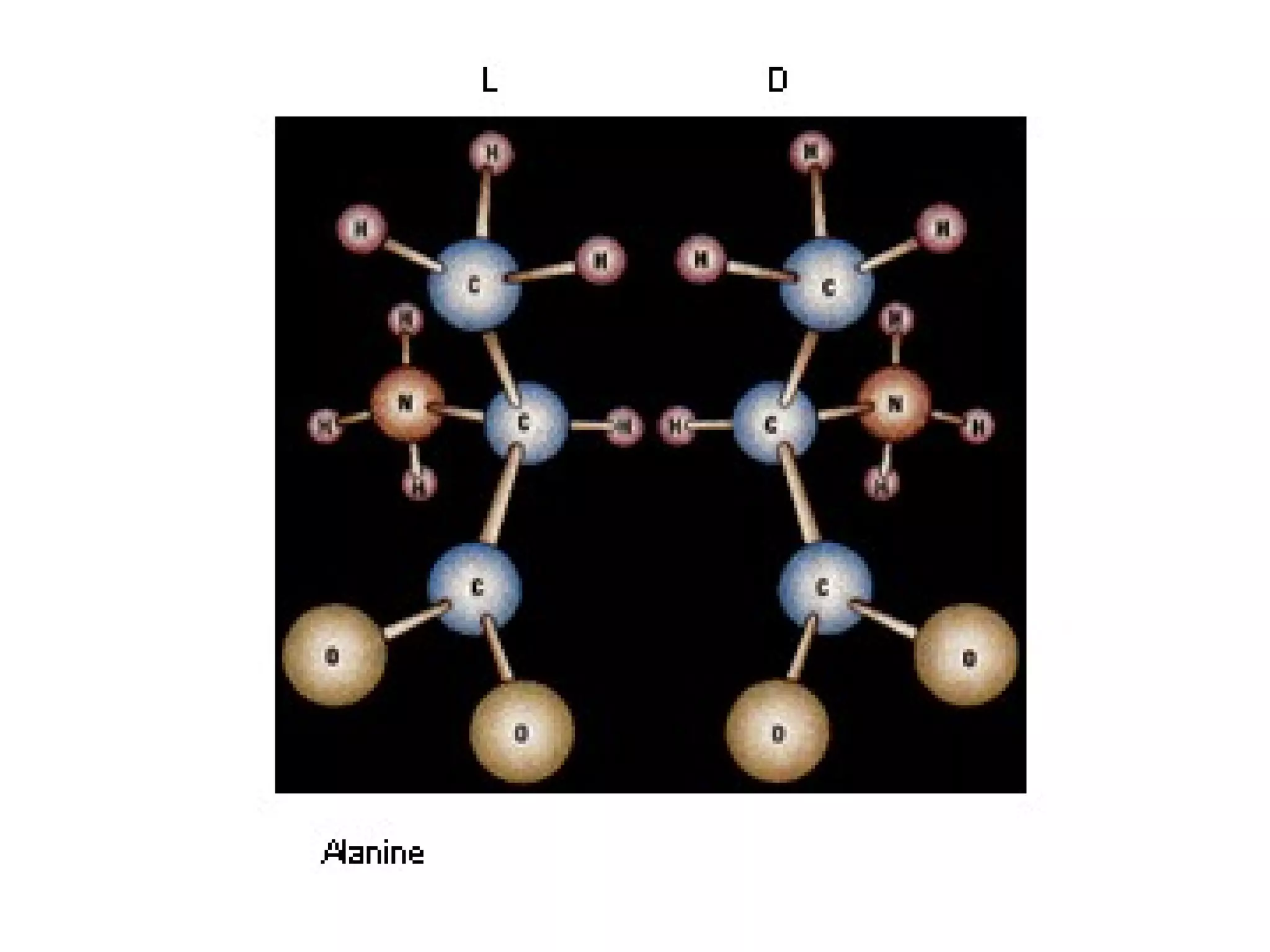
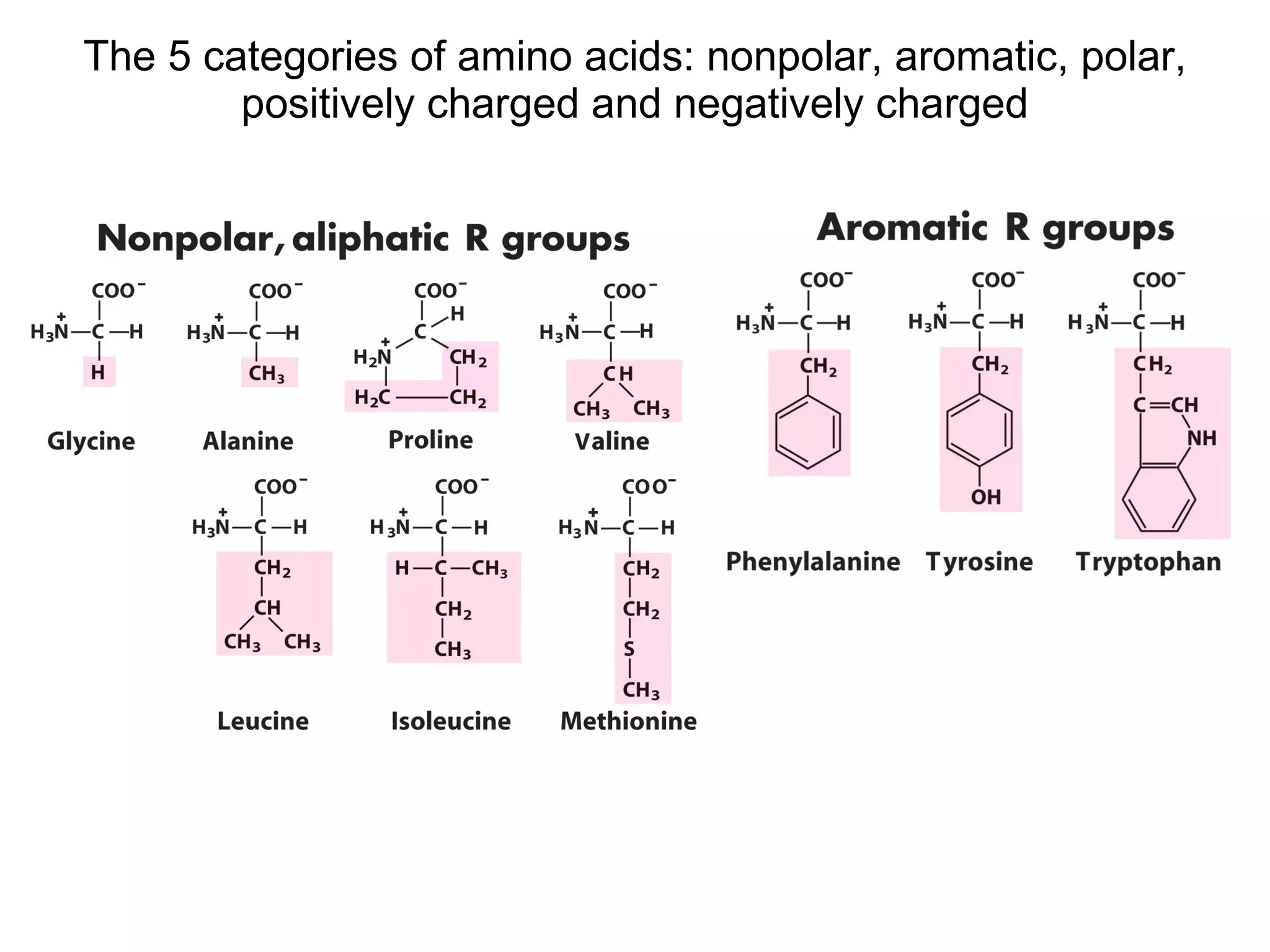
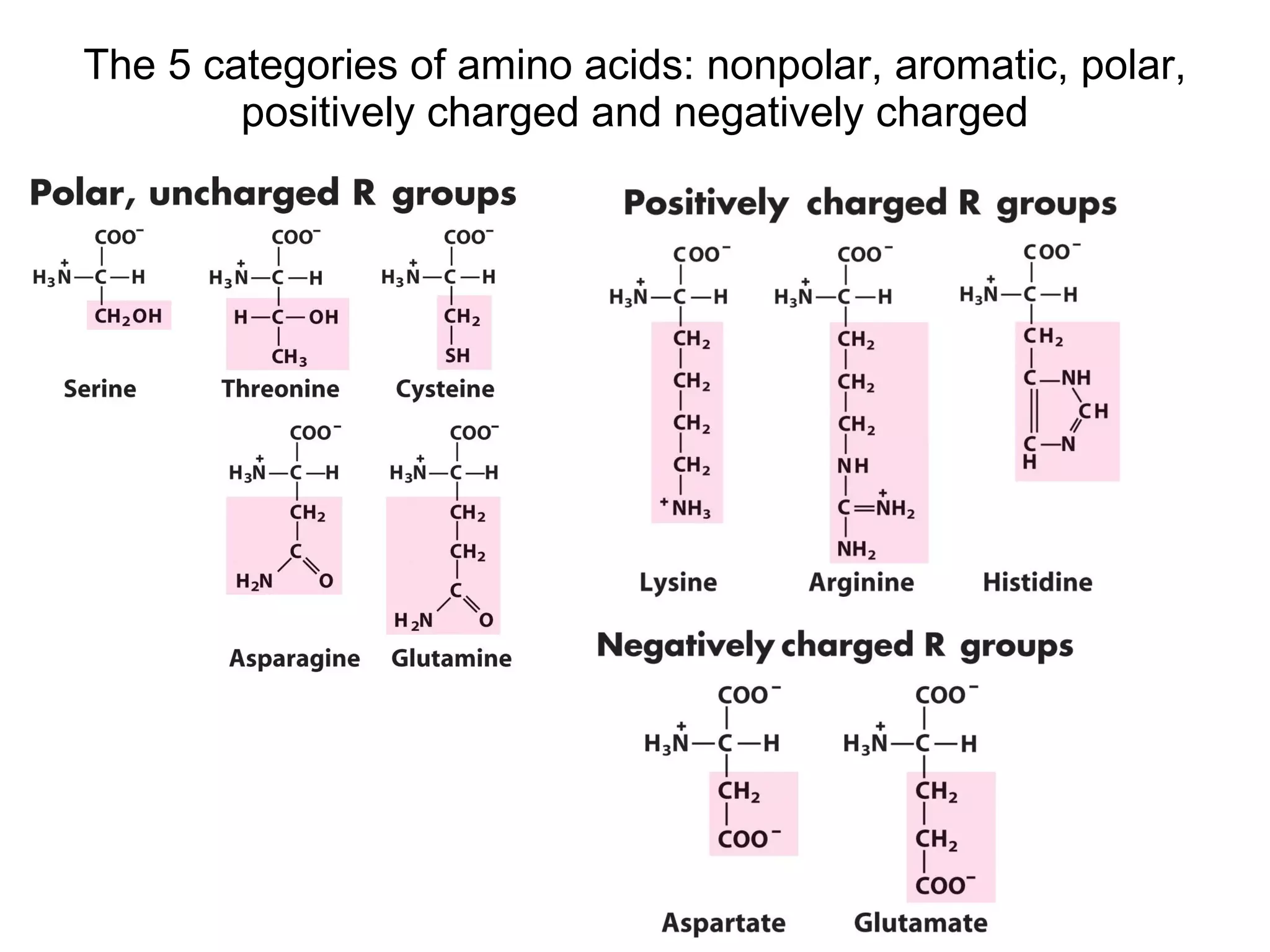
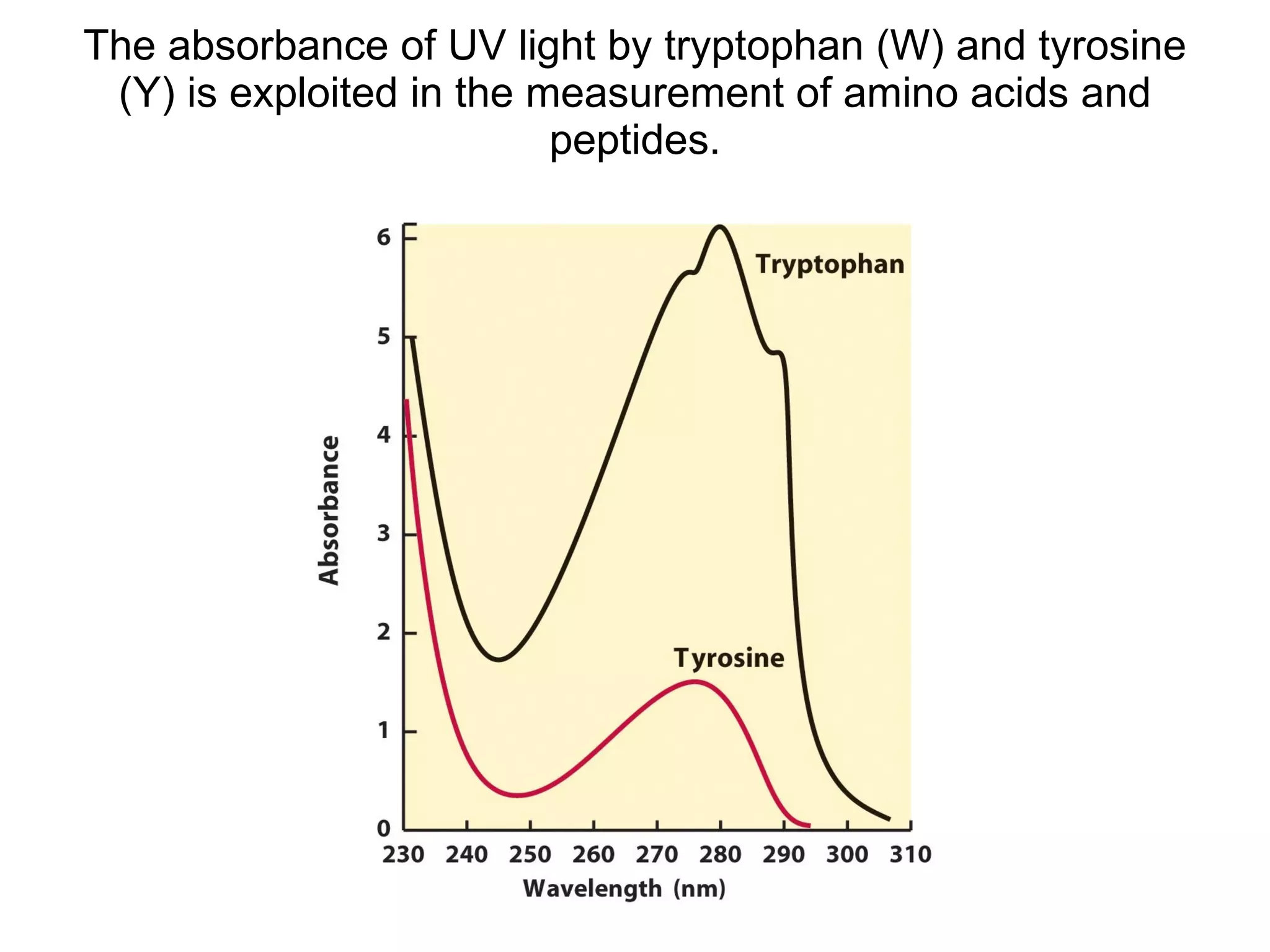
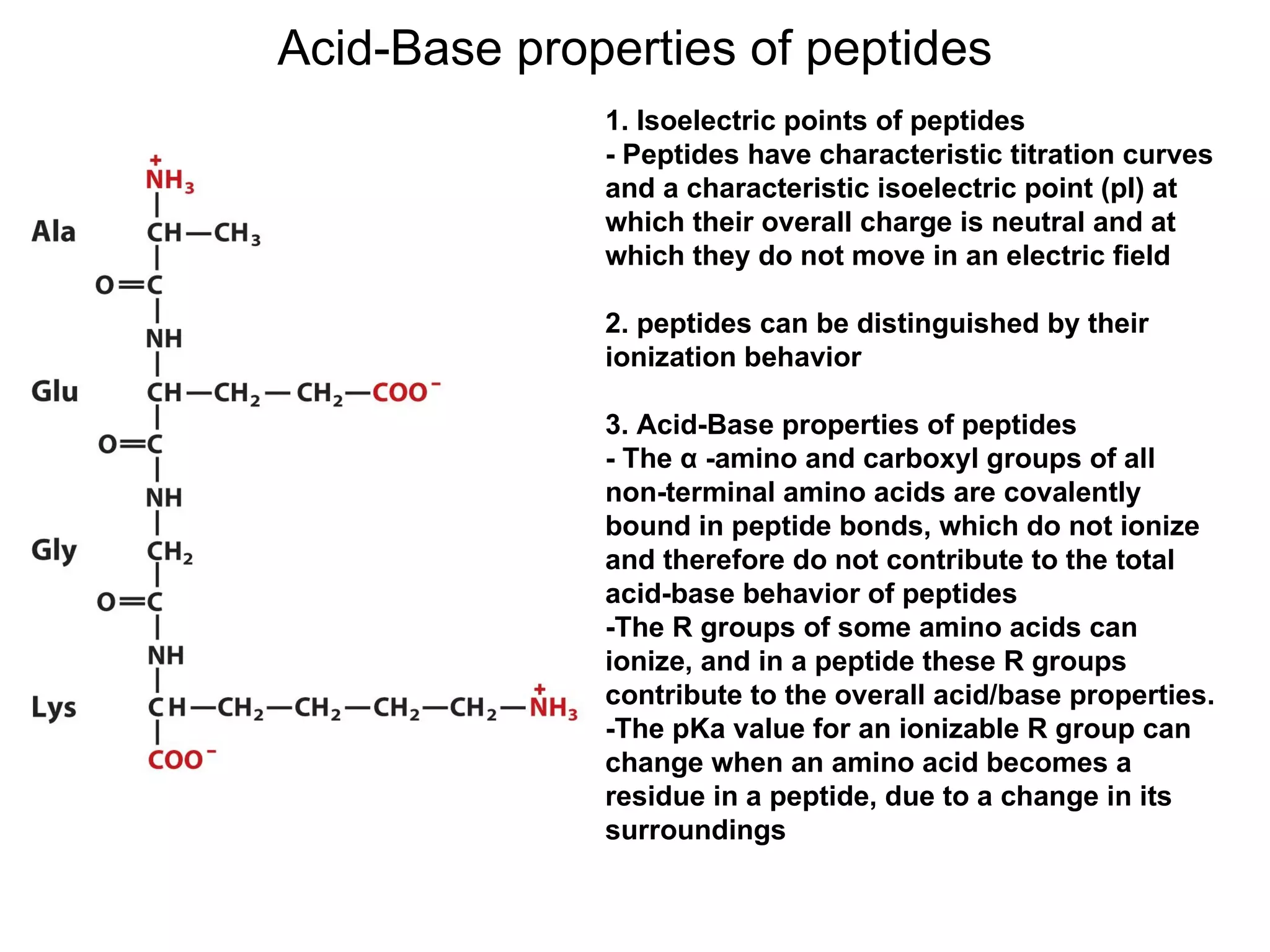
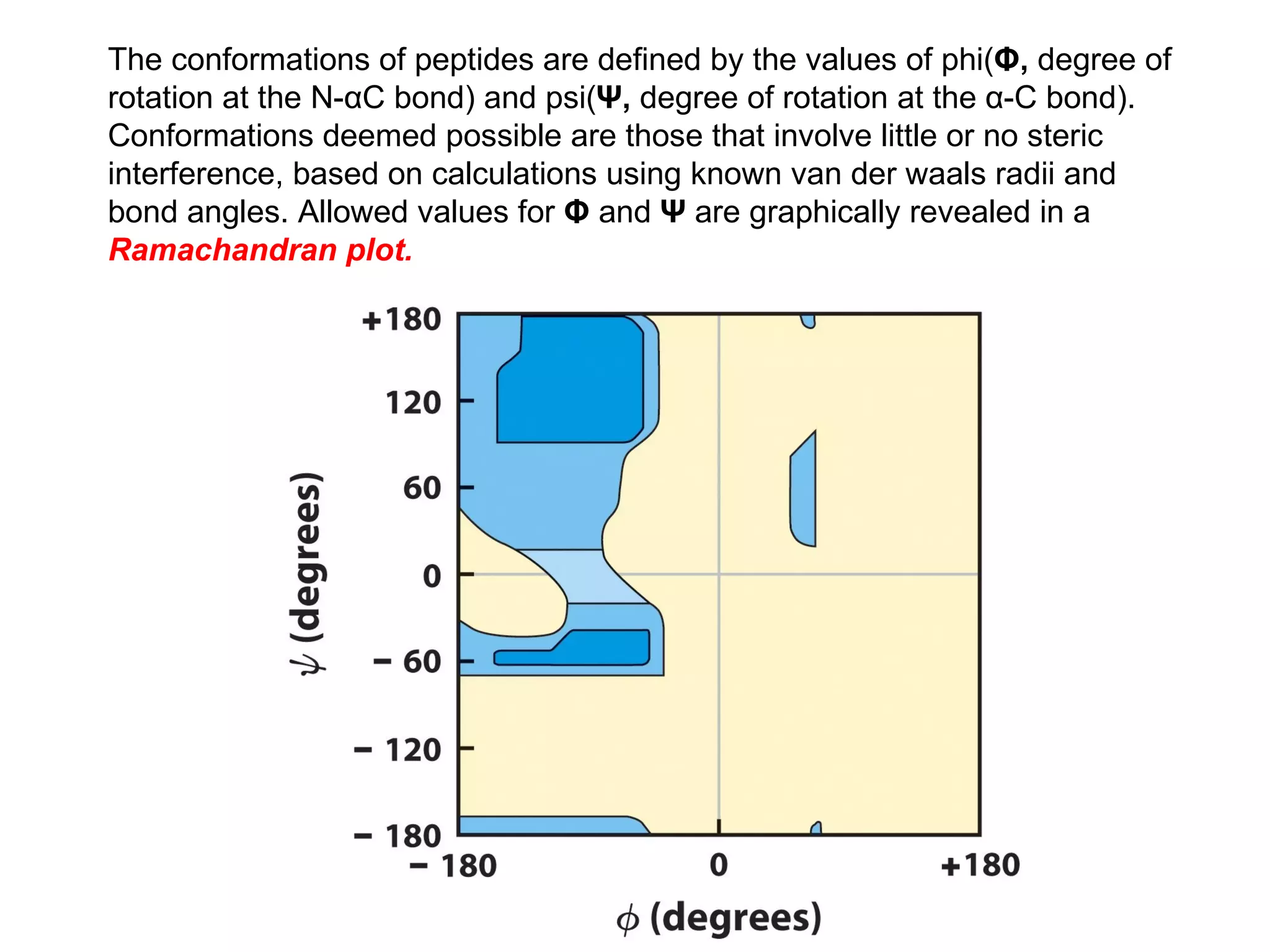
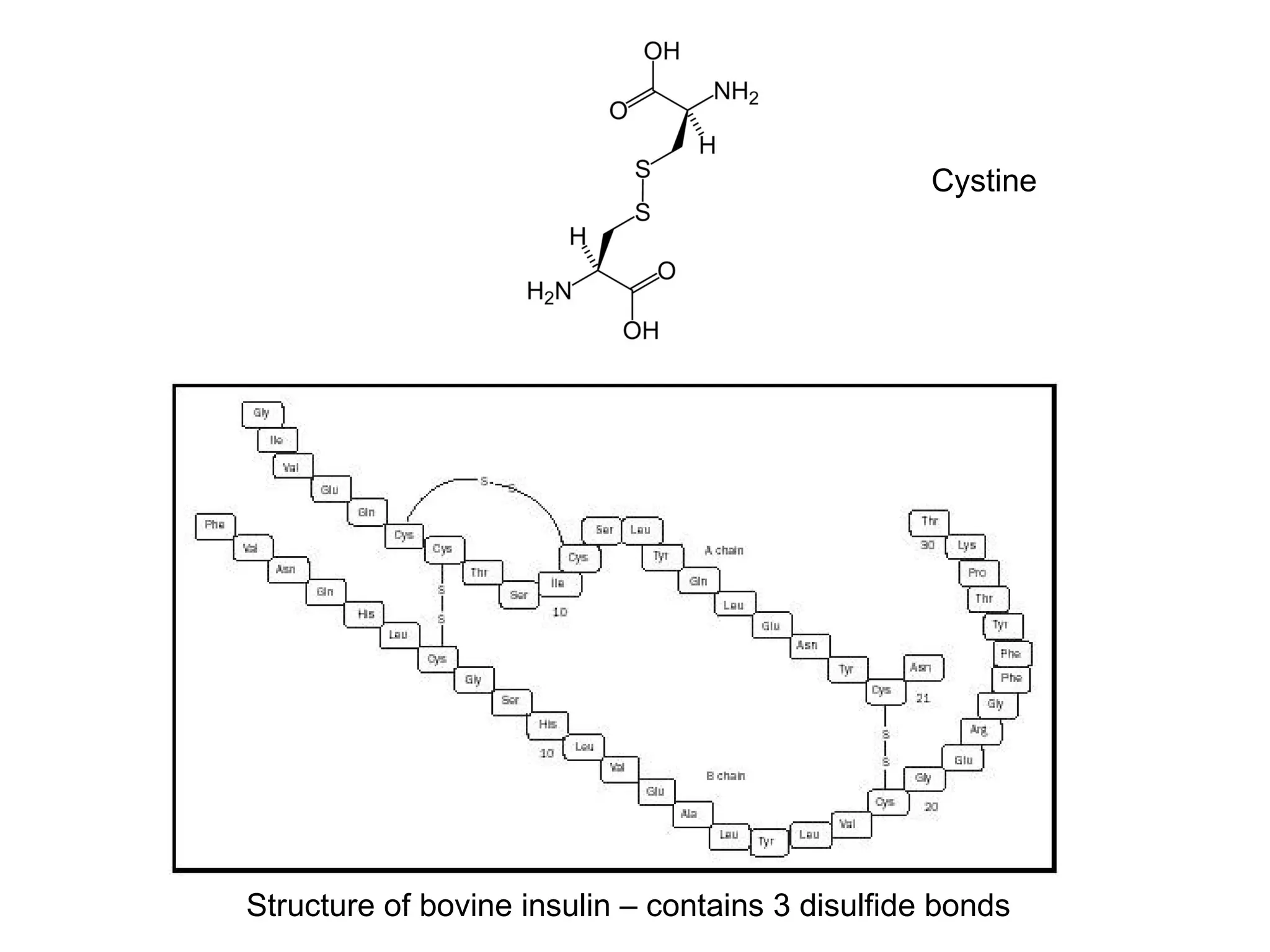
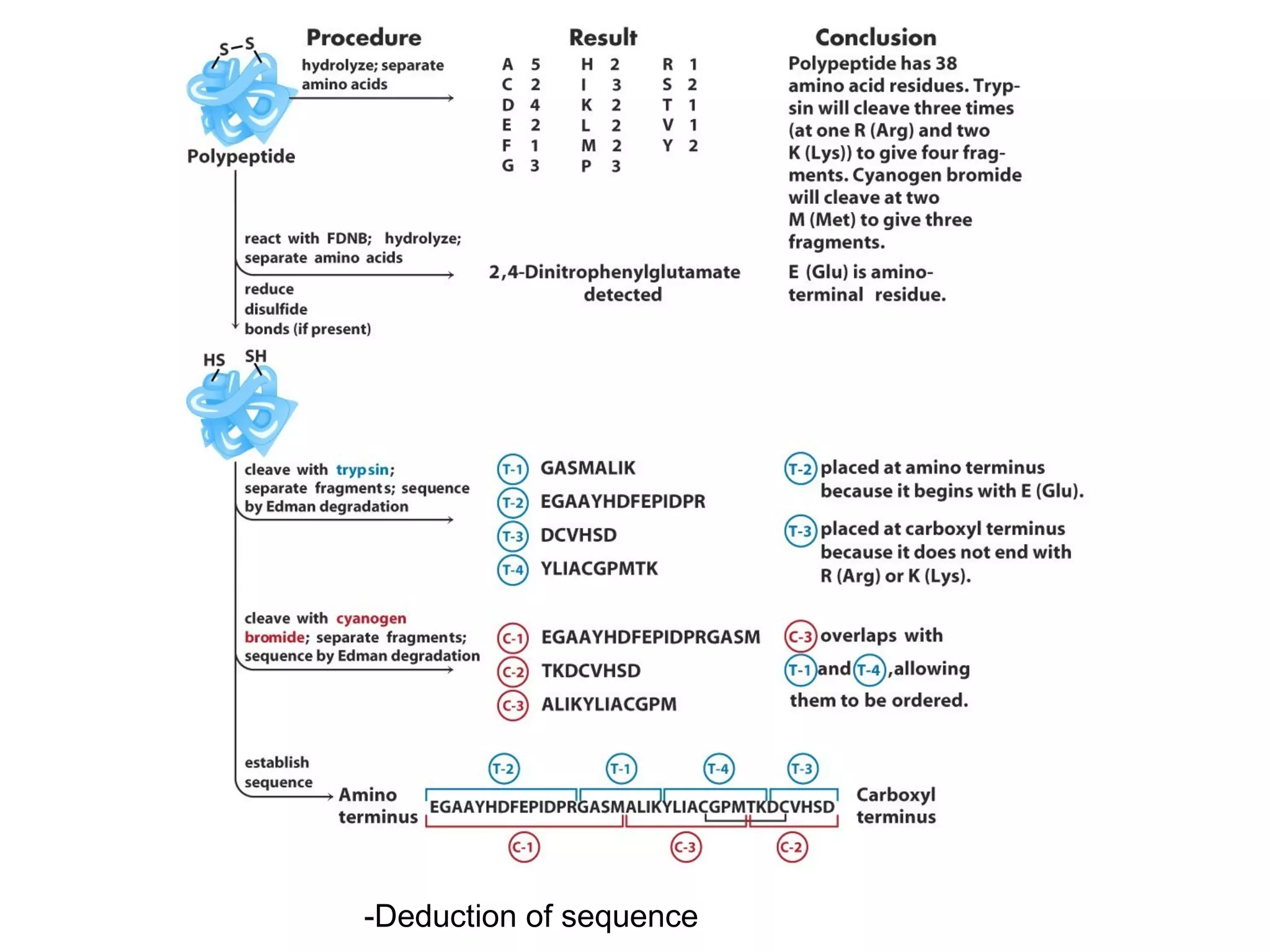
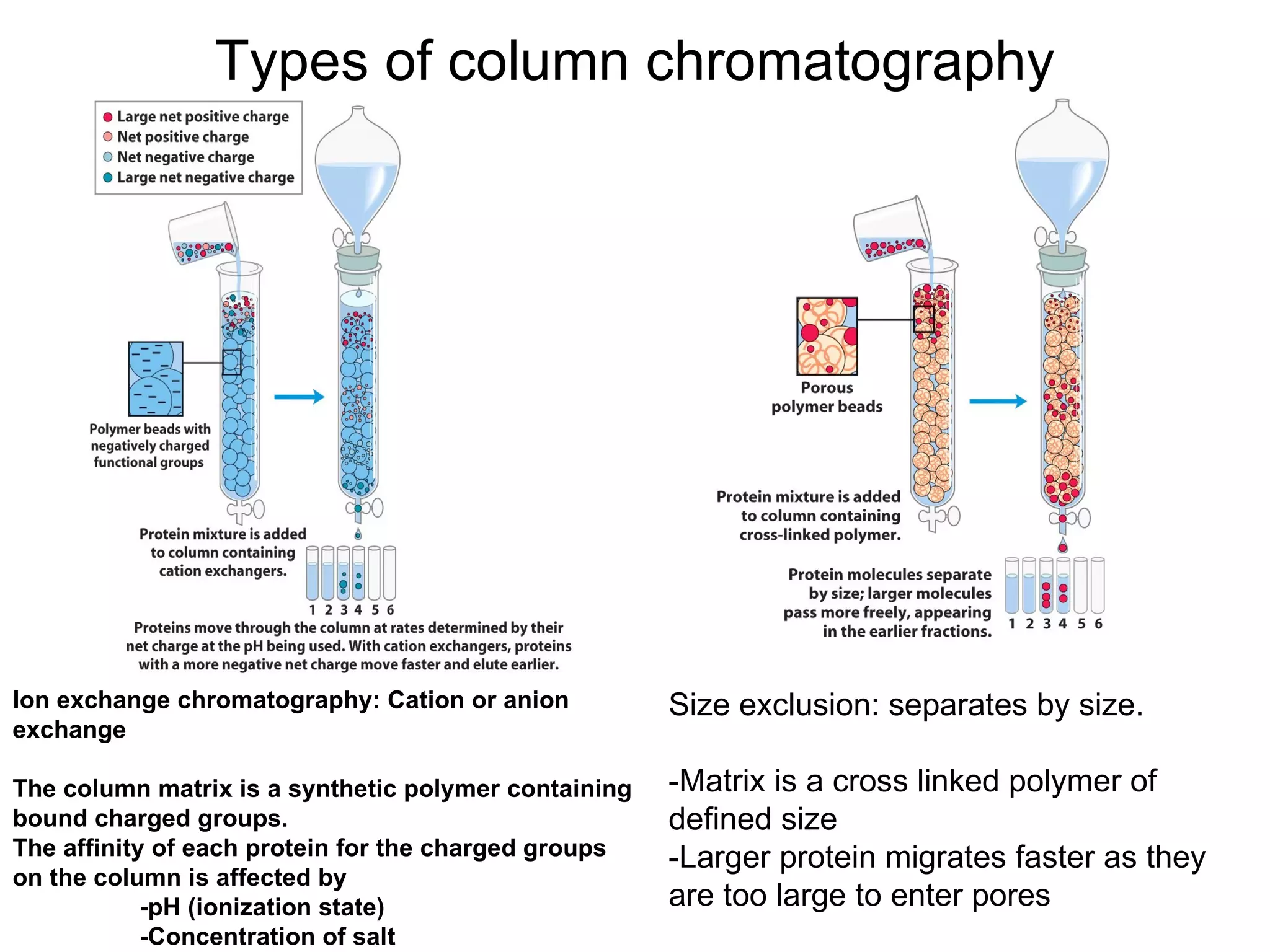
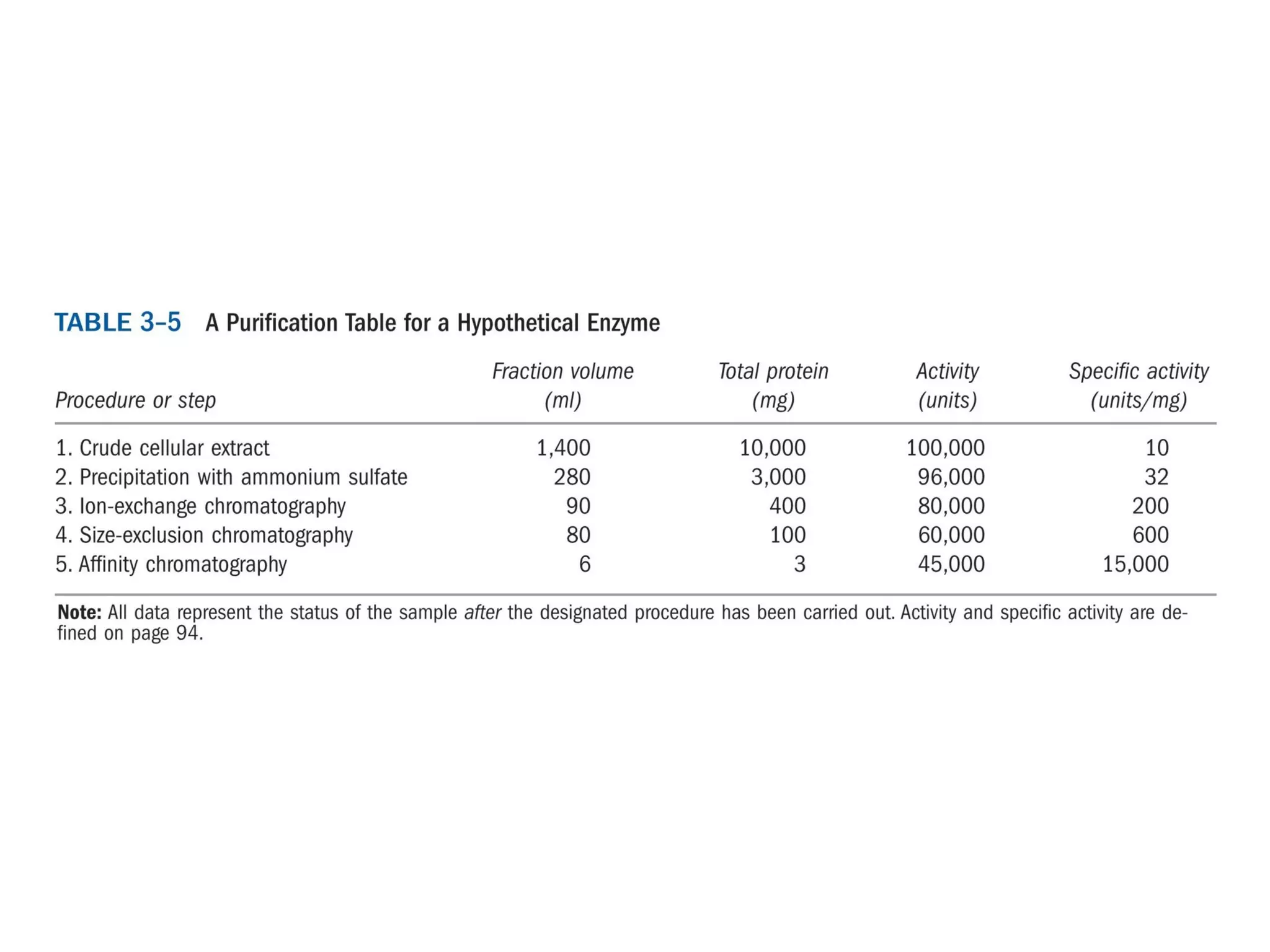
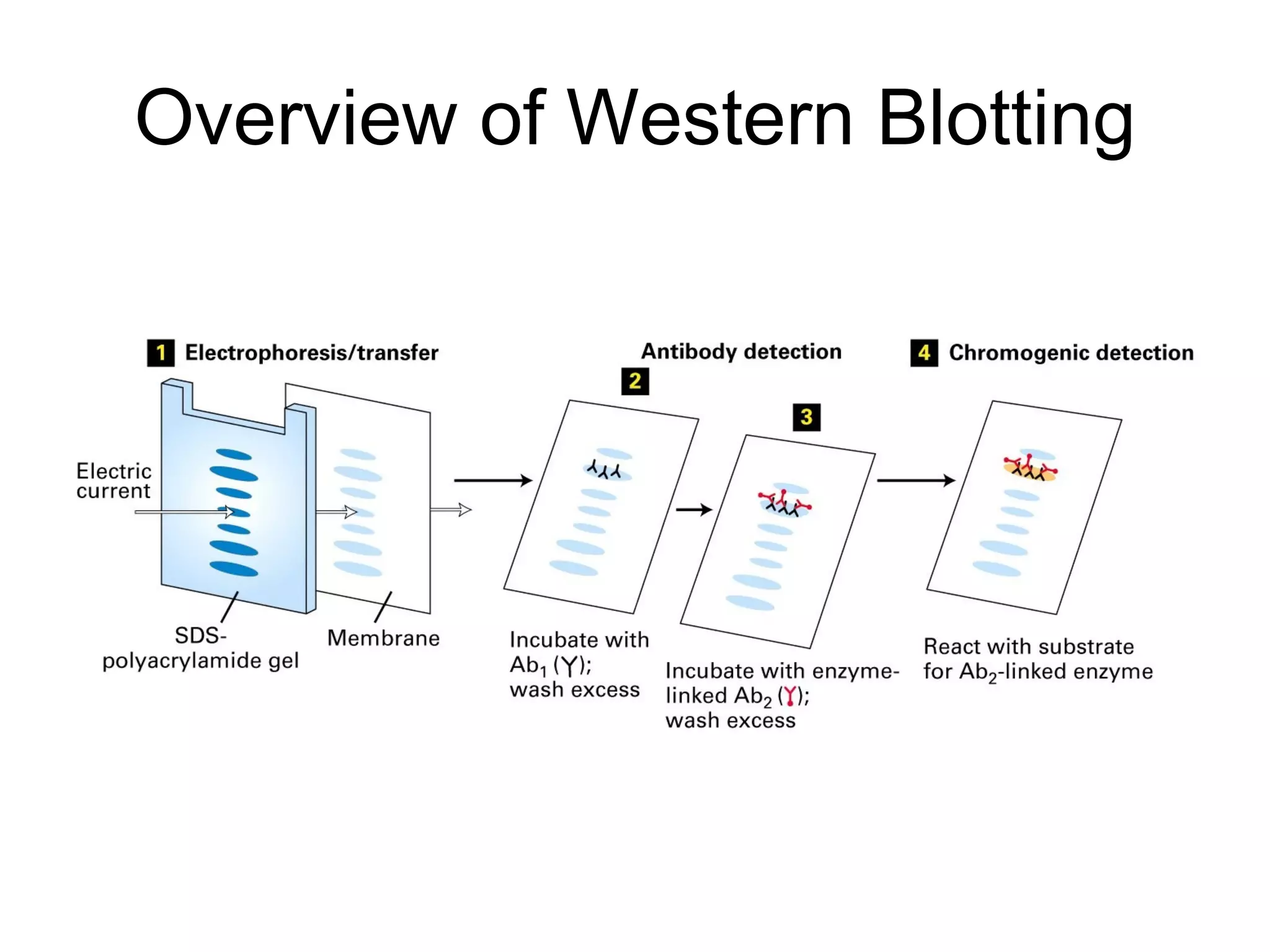
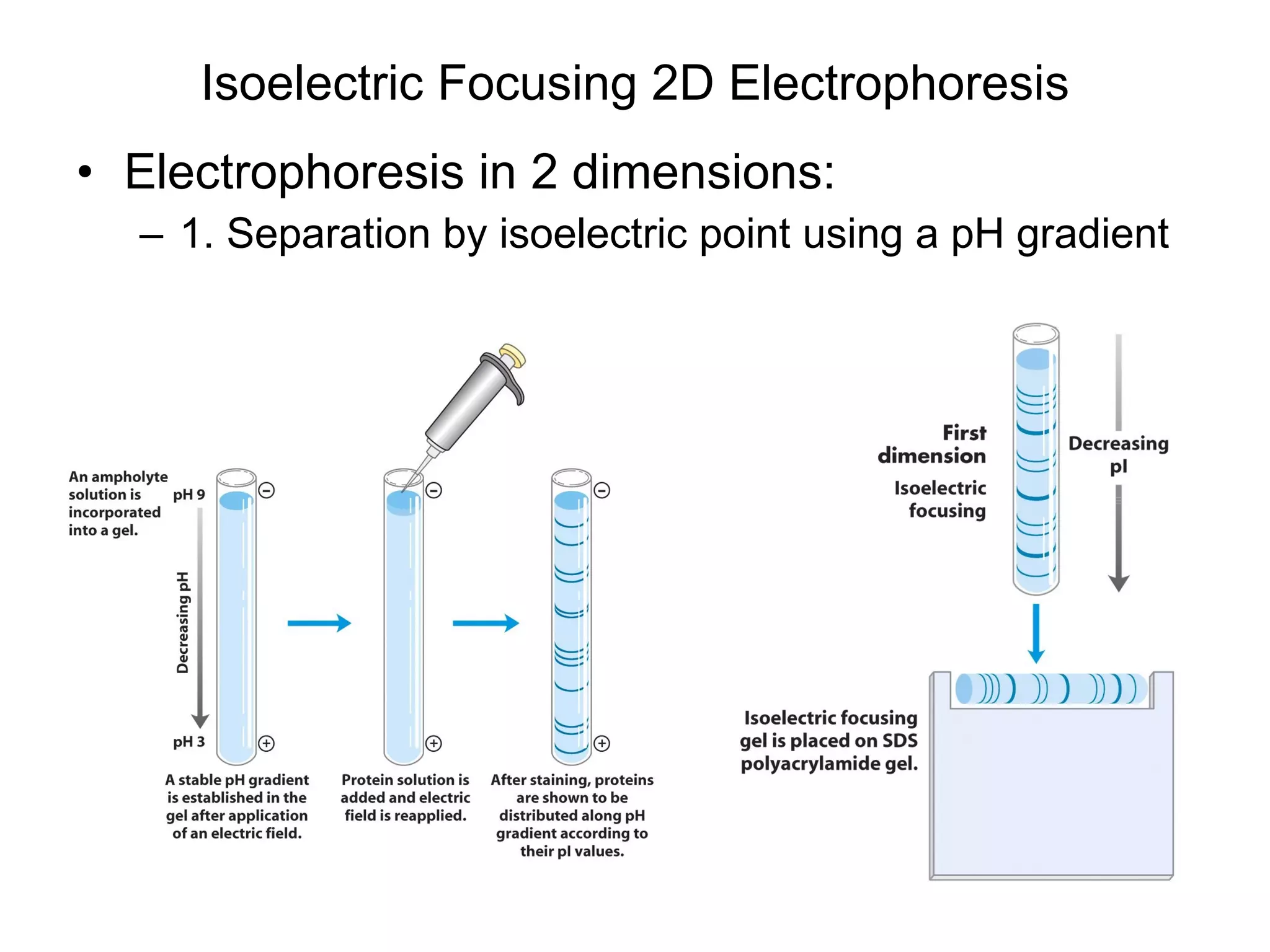
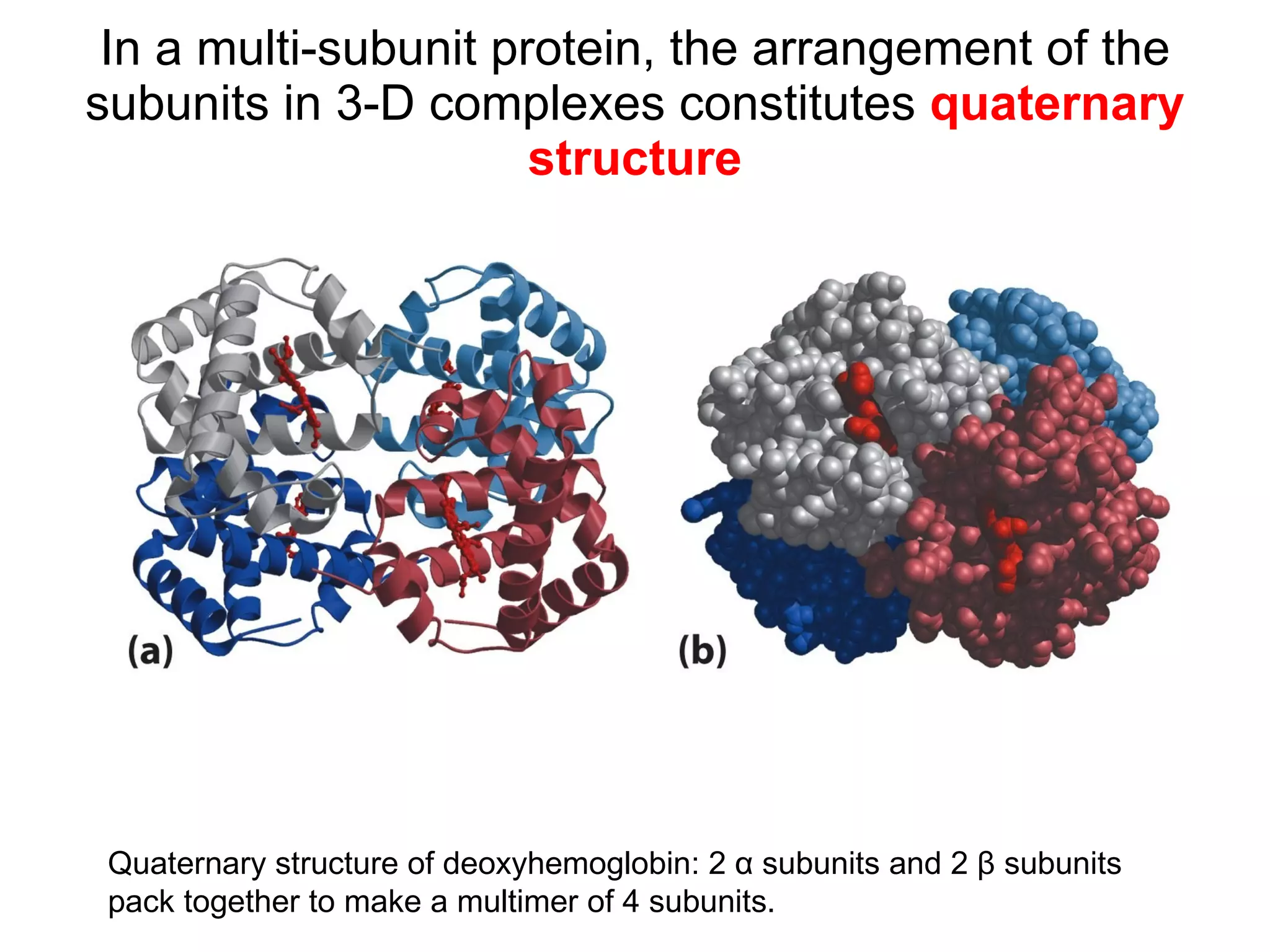
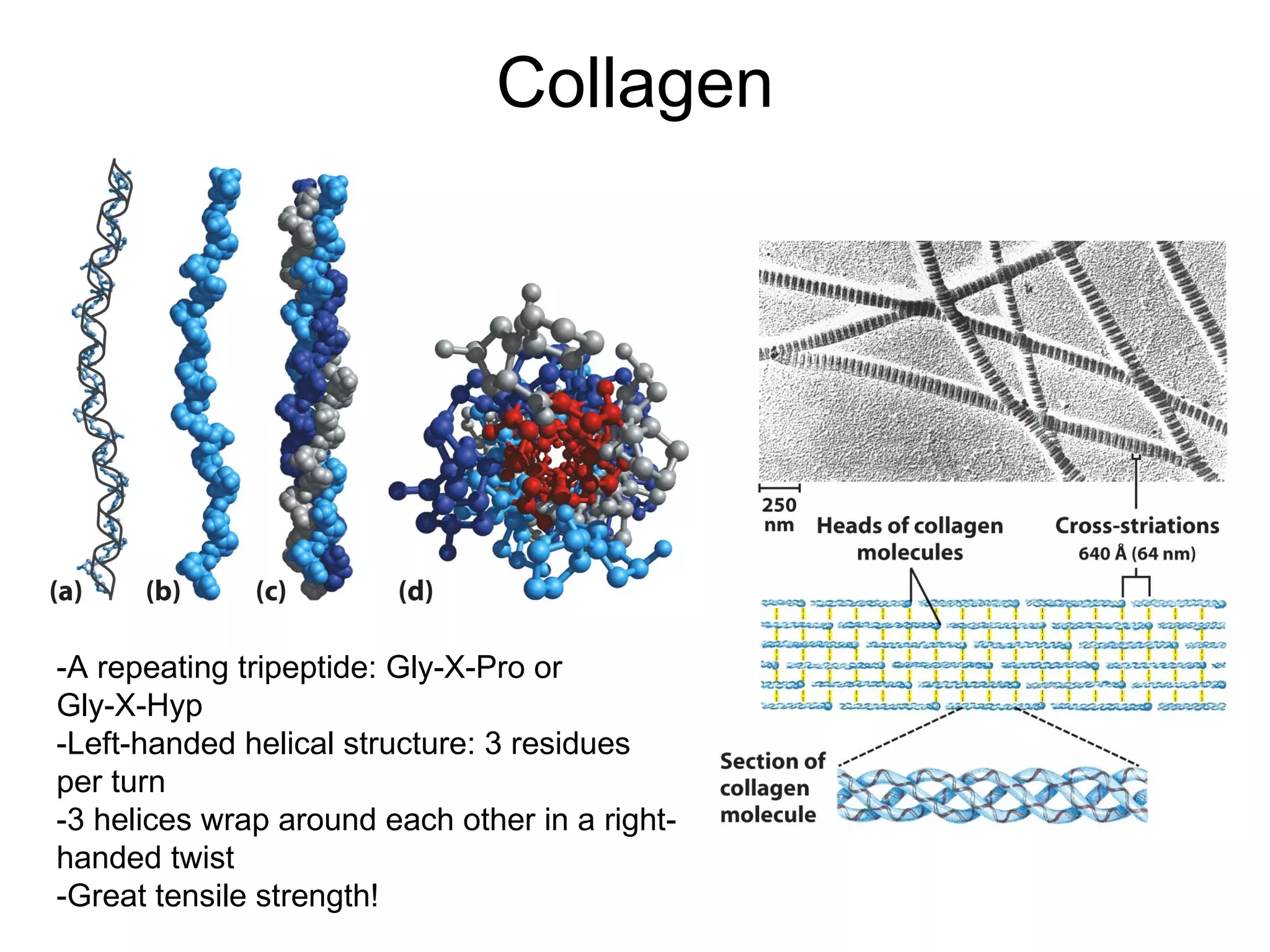
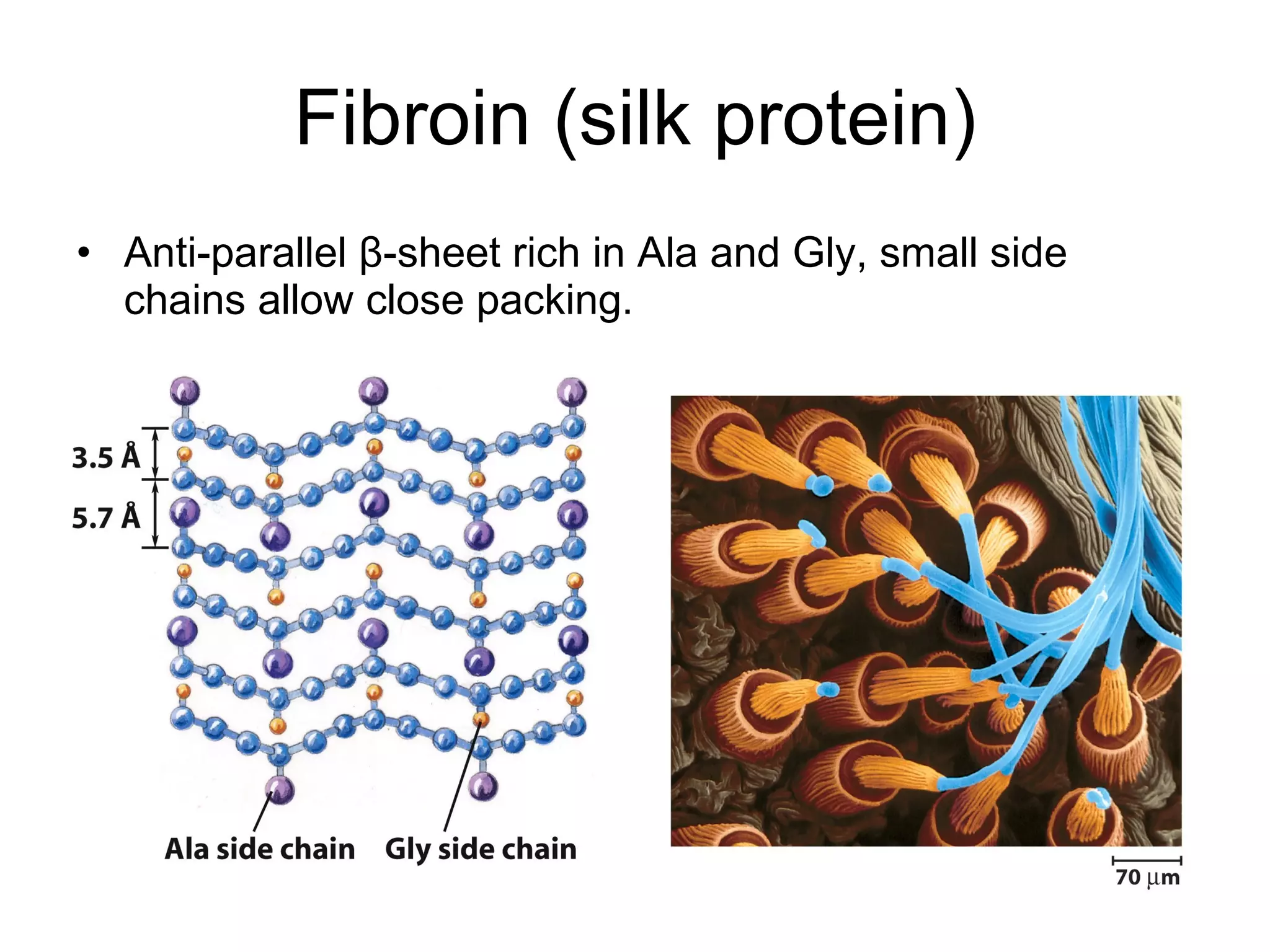
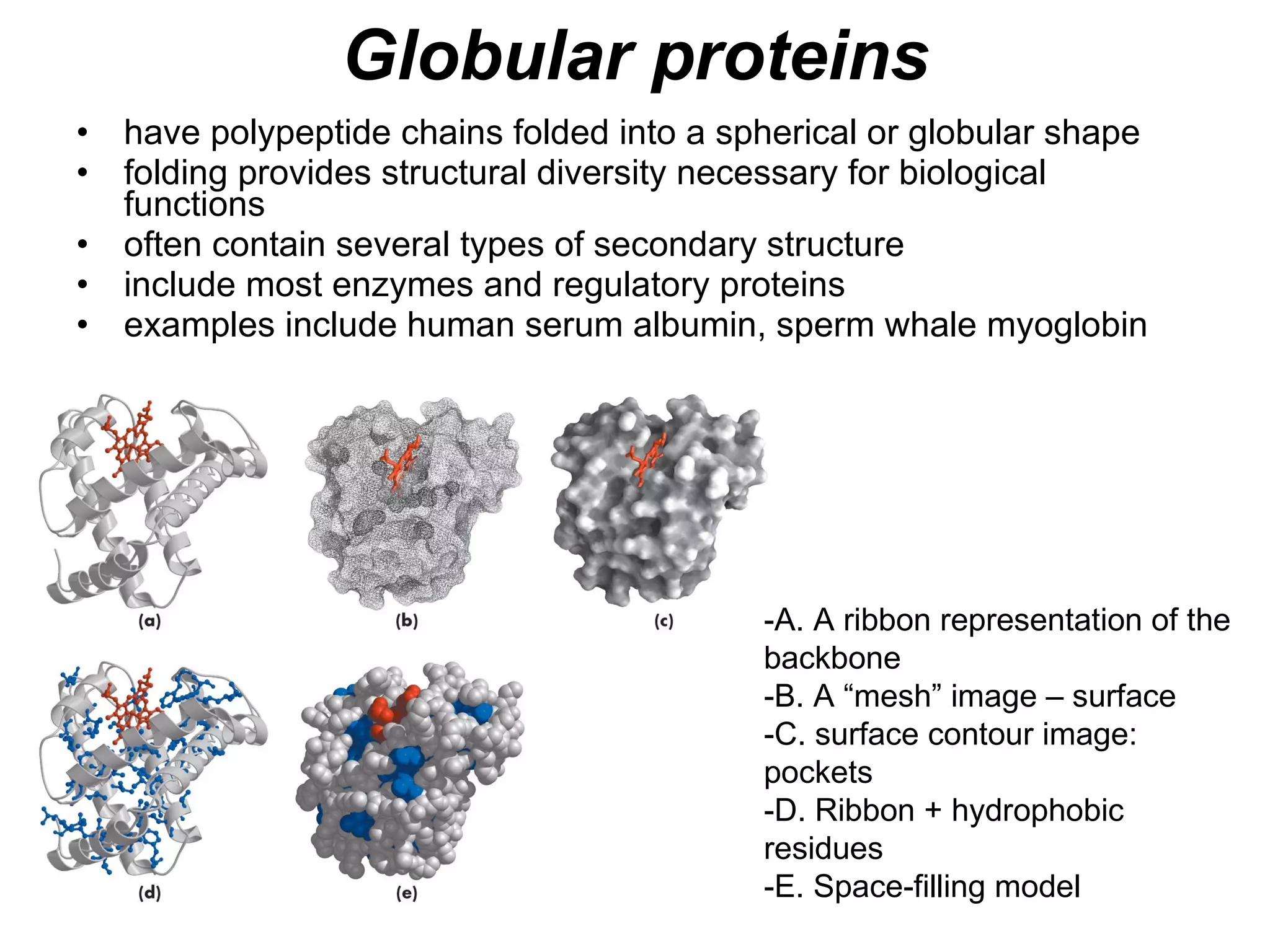
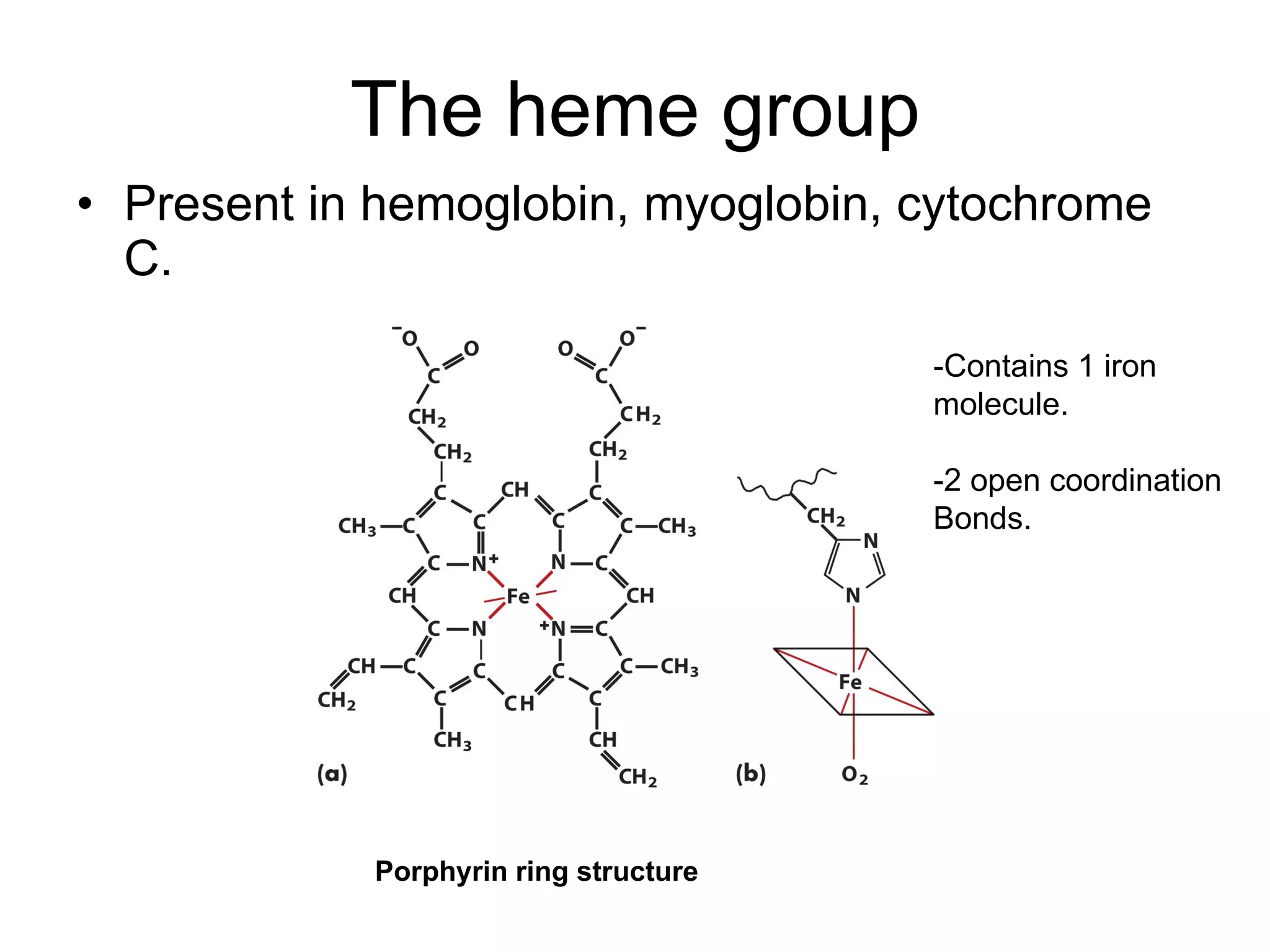
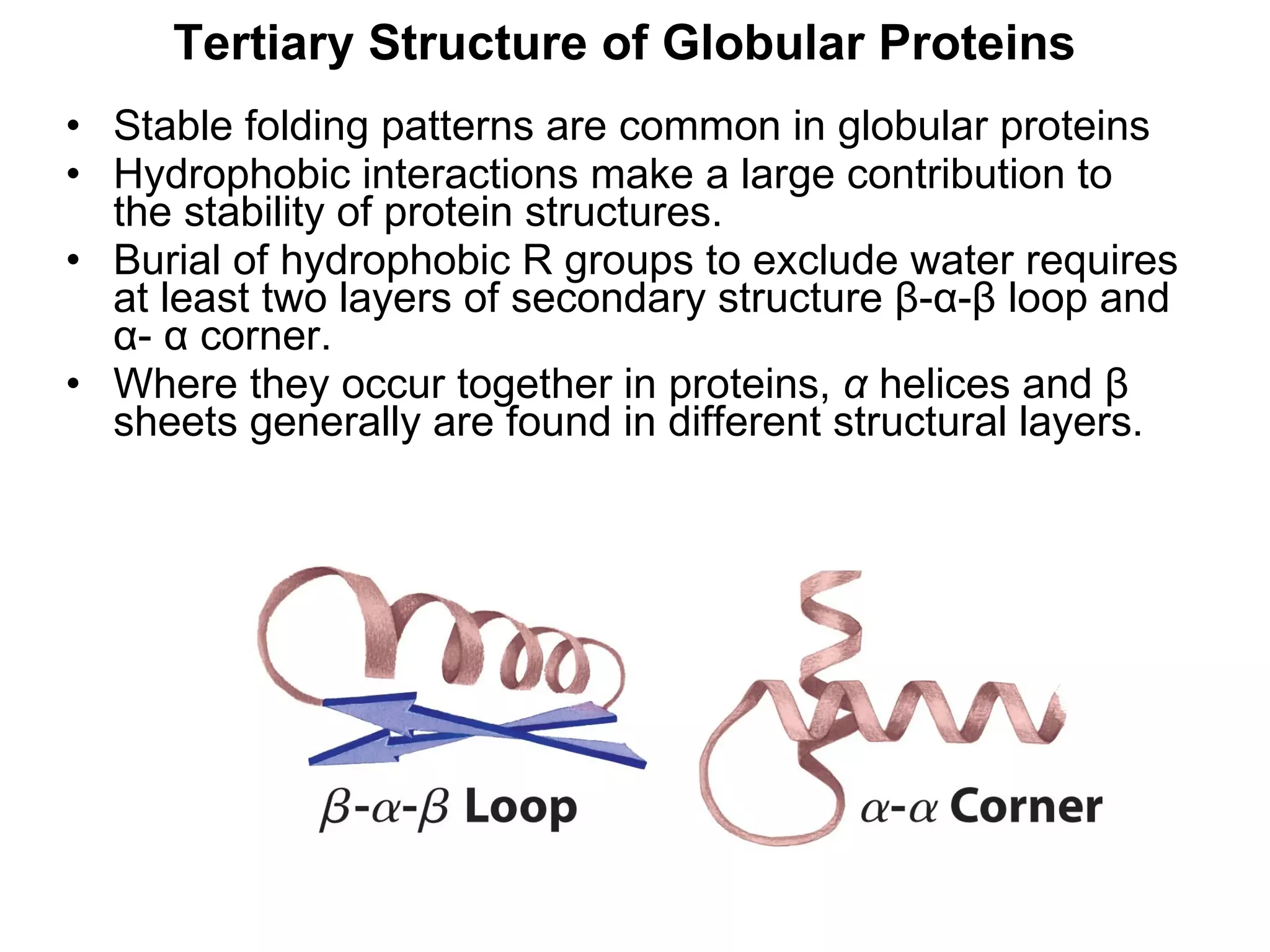
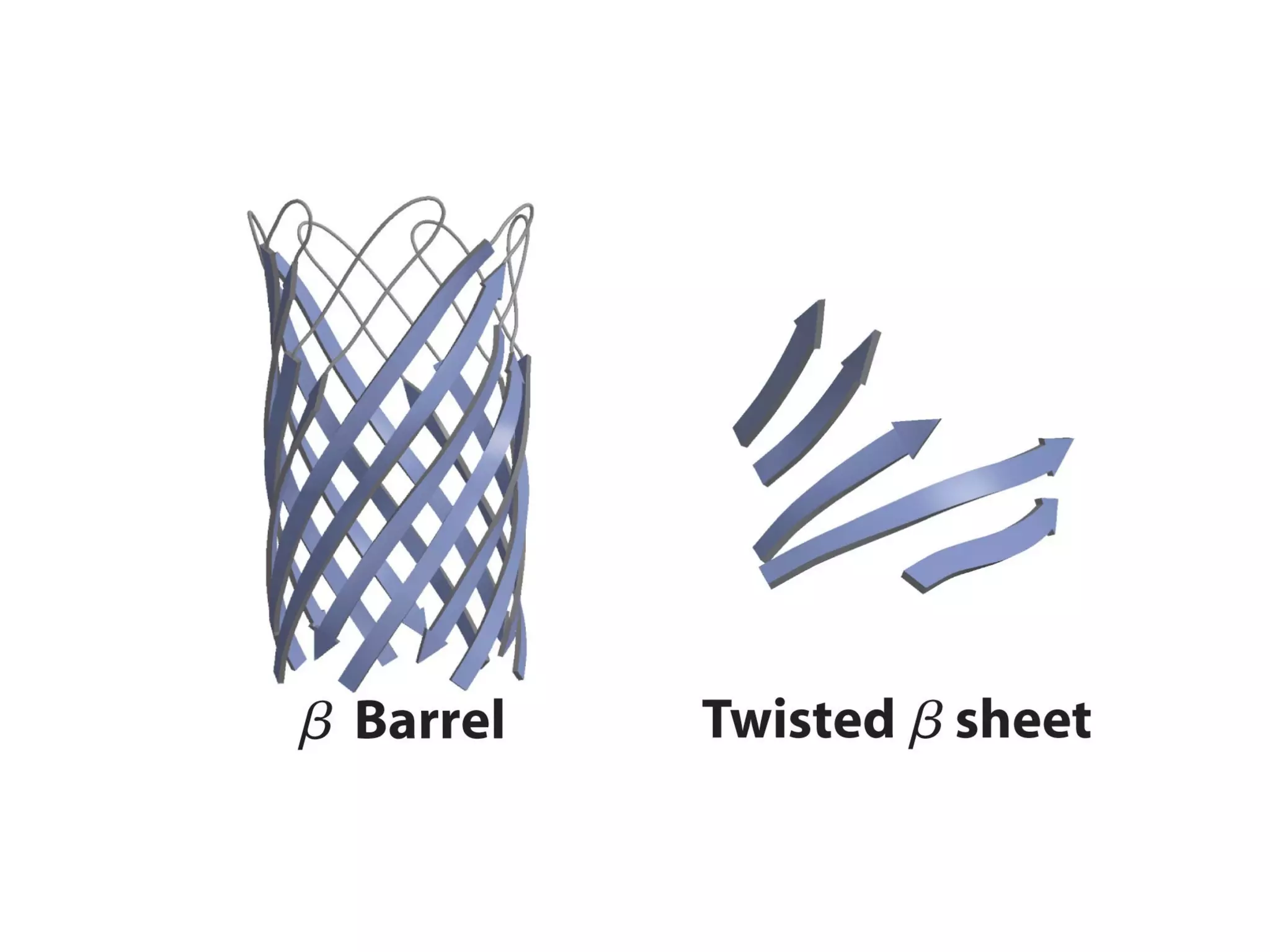
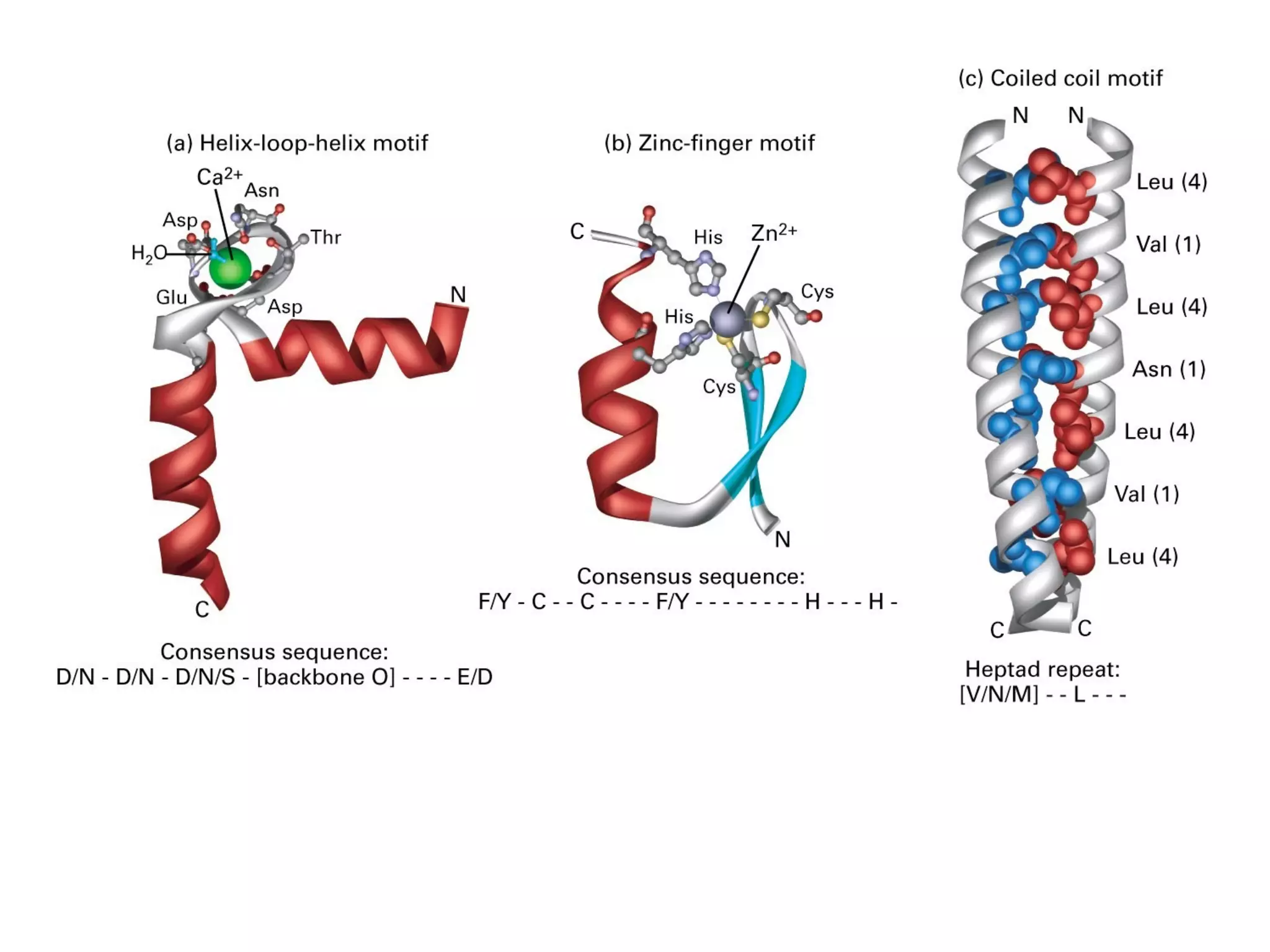
The document summarizes key aspects of amino acids and protein structure in 3 paragraphs or less:
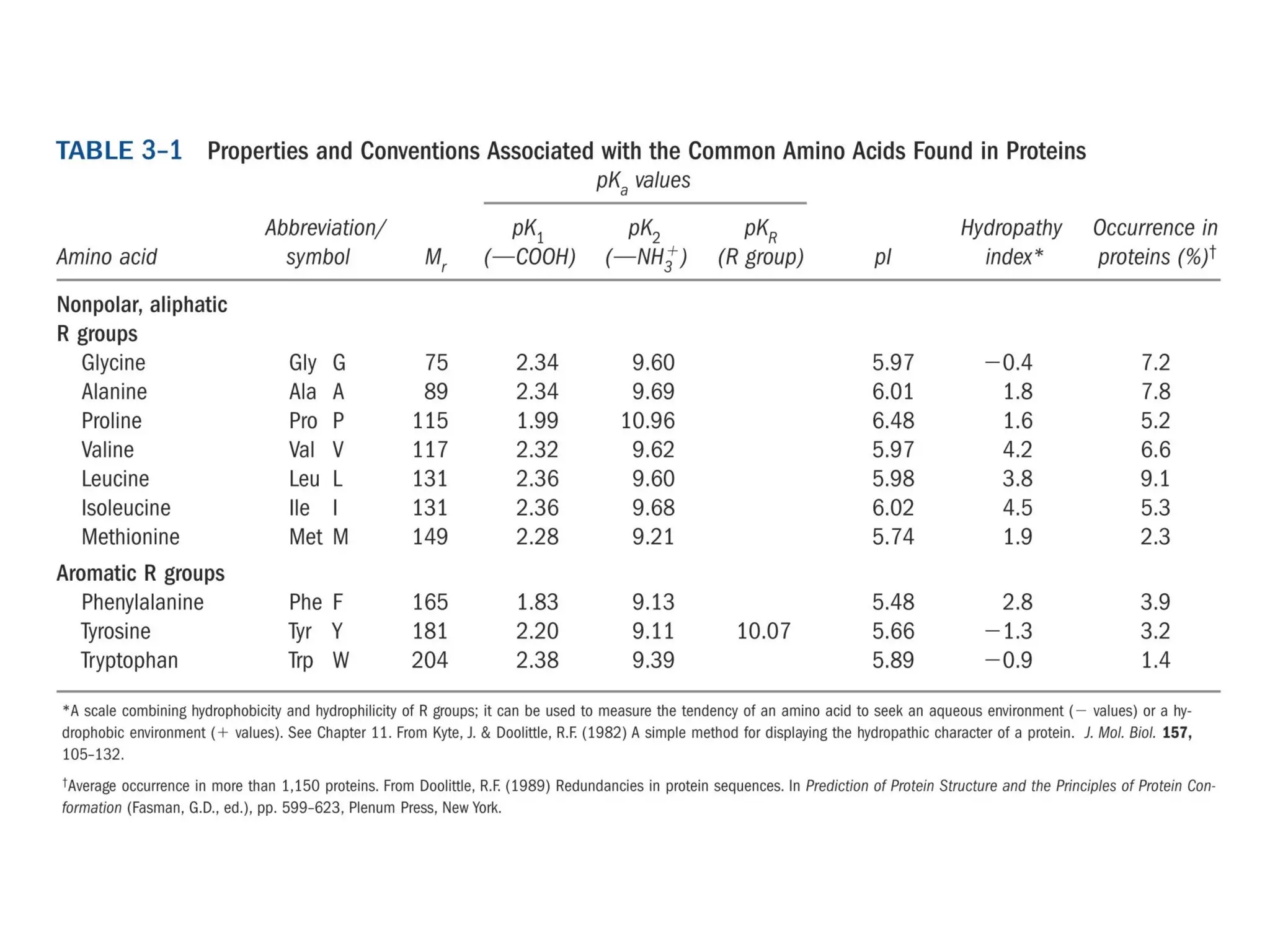
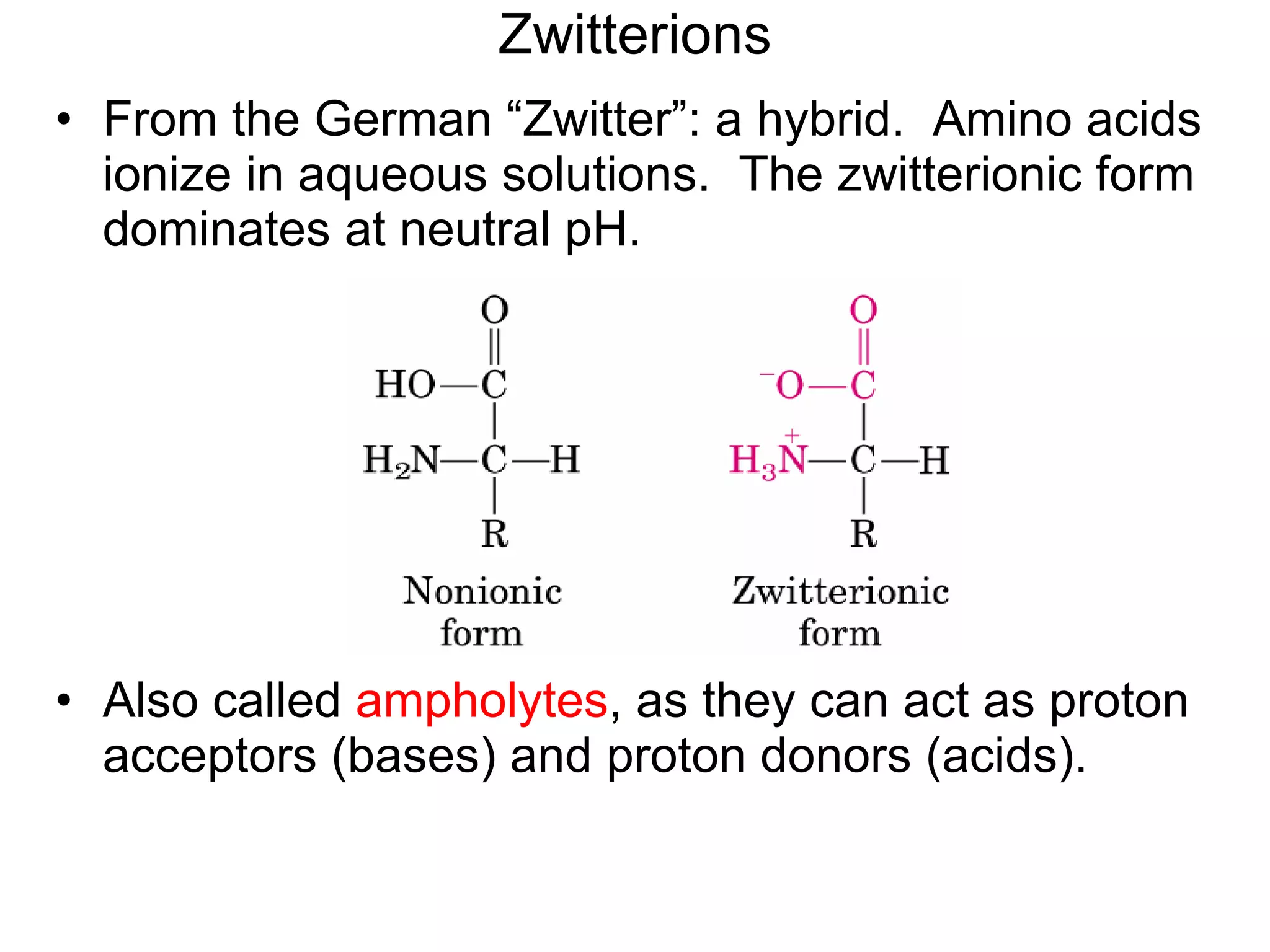
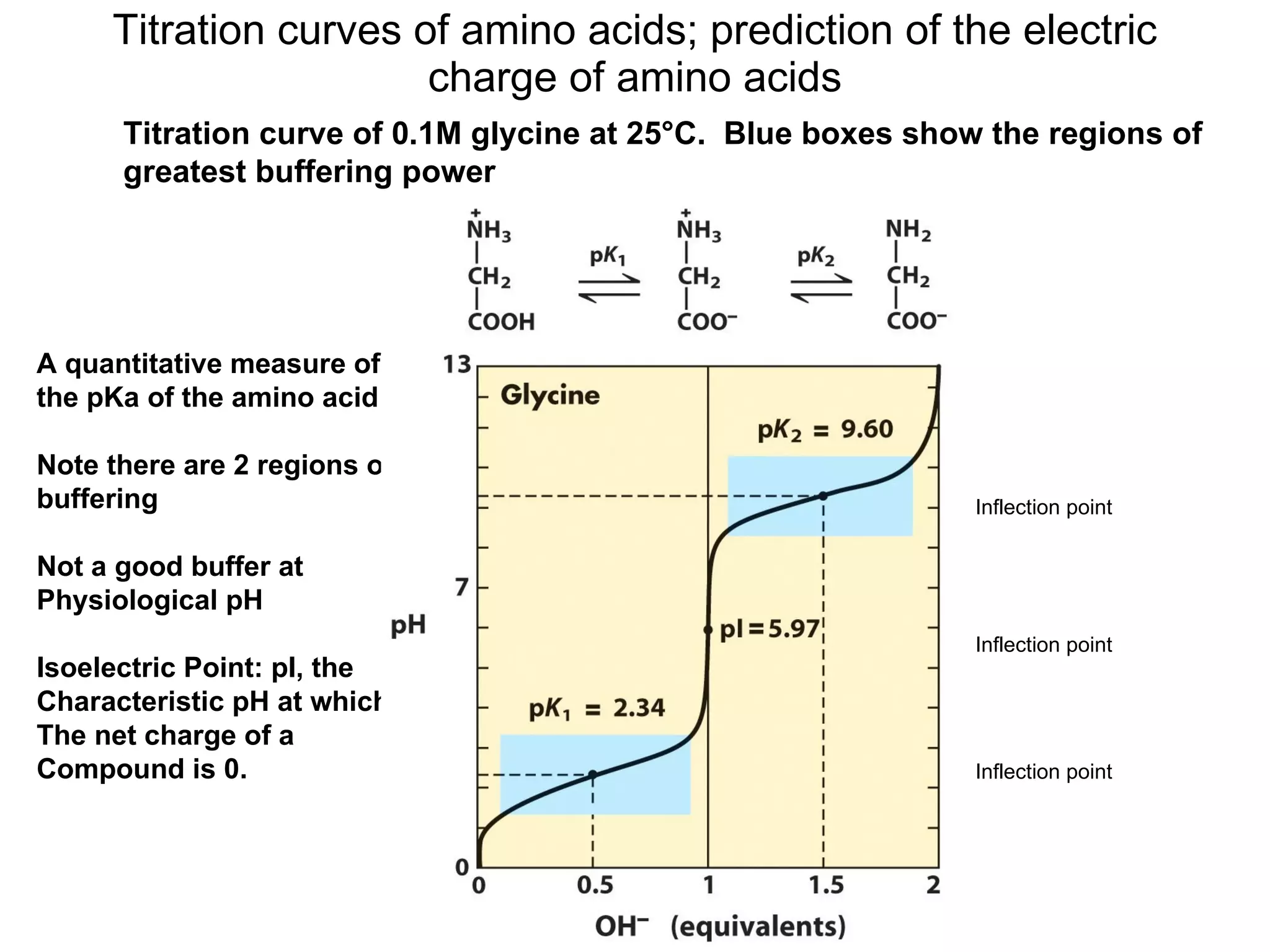
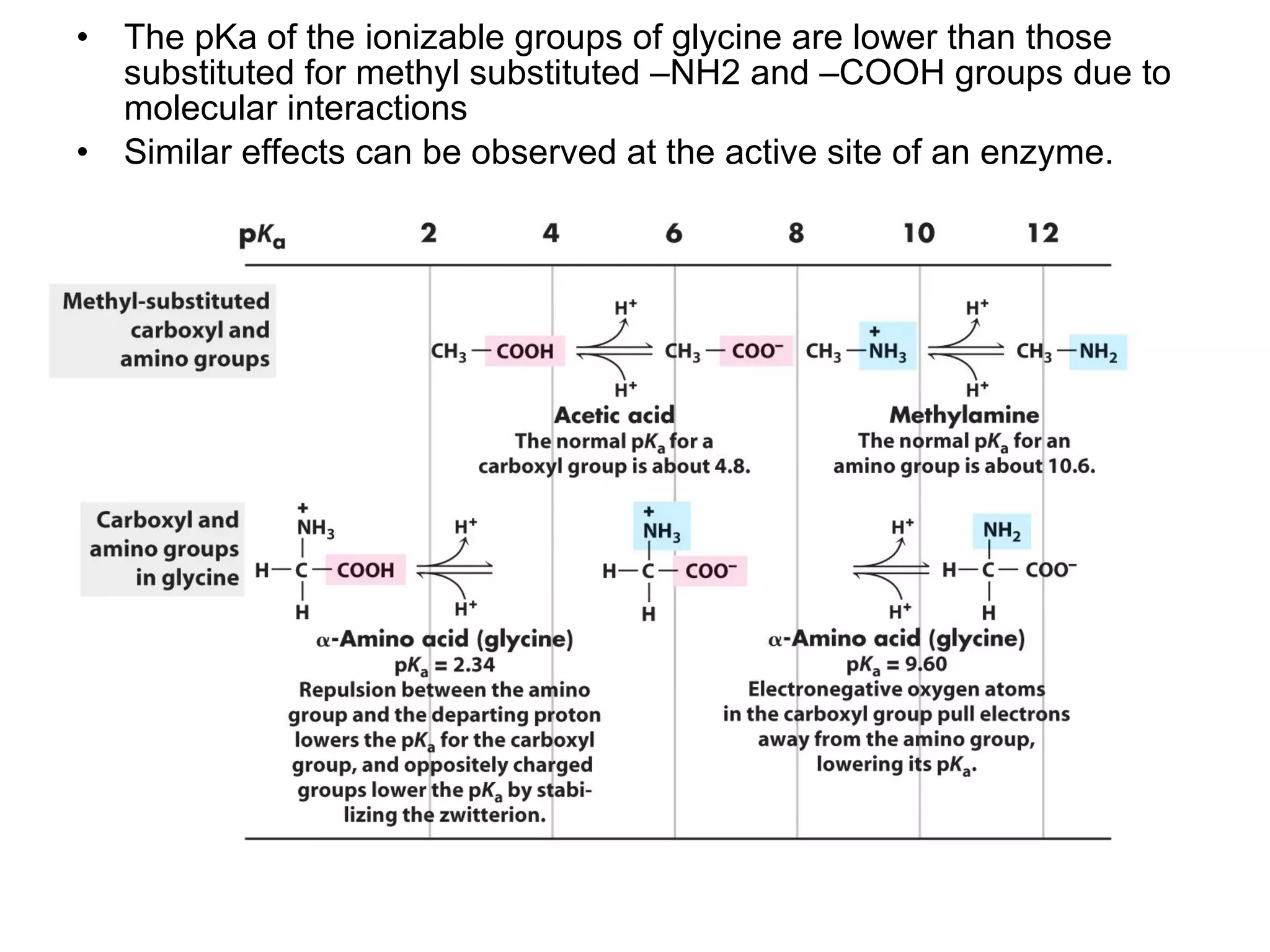
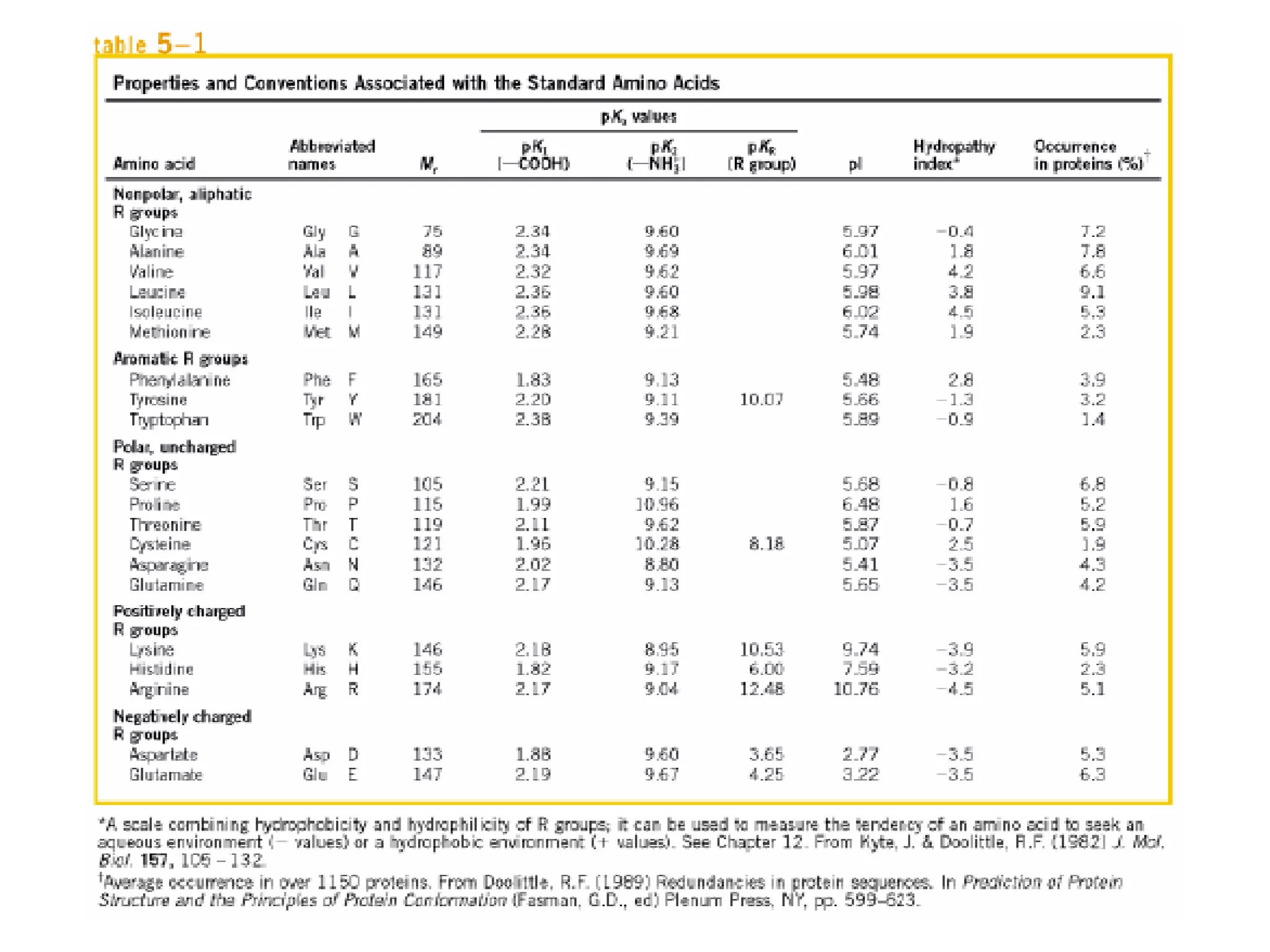
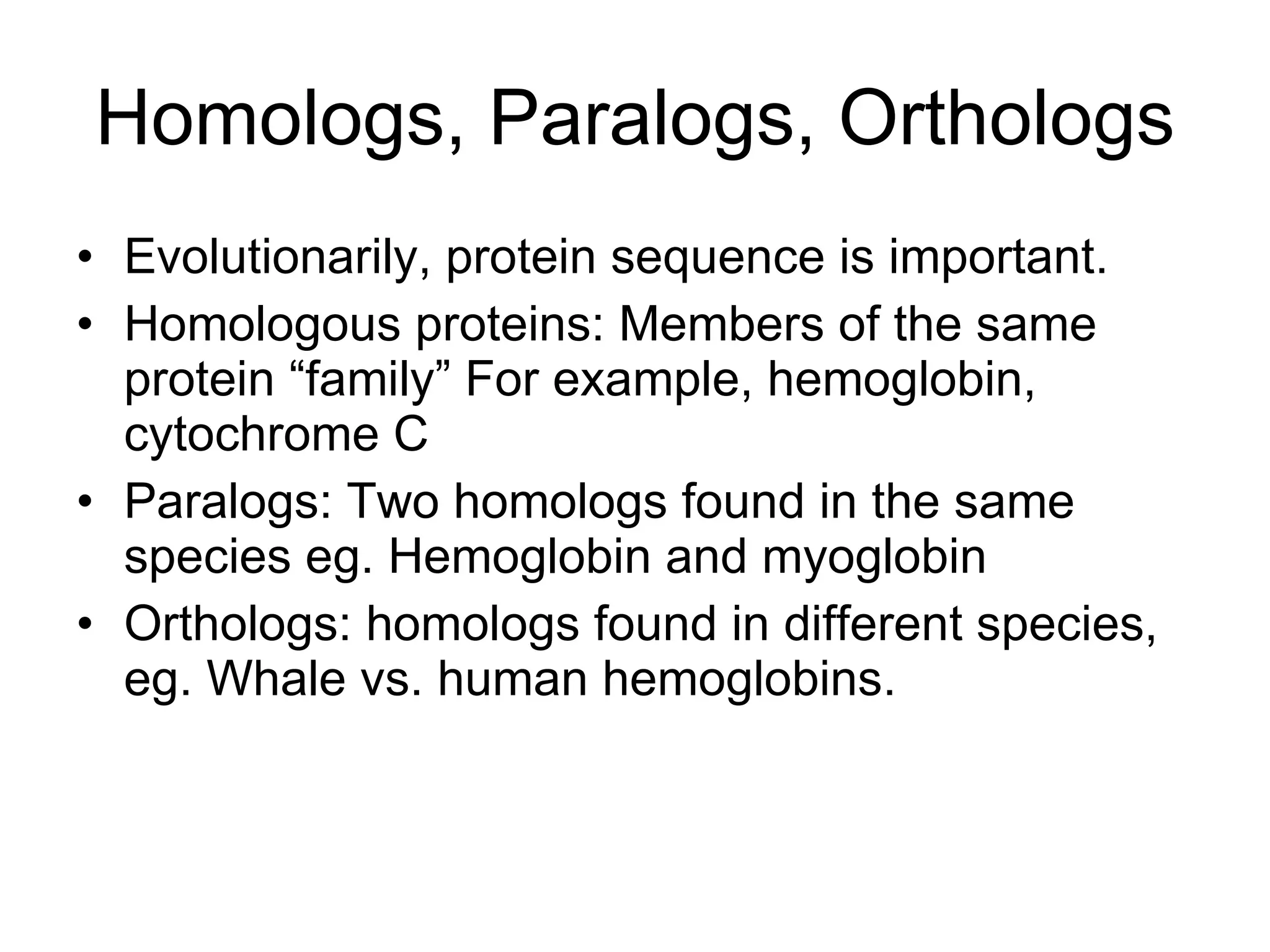
Amino acids are the building blocks of proteins. They contain common structural features and exist in L- and D-forms. In proteins, amino acids are exclusively in the L-conformation. Amino acids are classified based on the properties of their side chains into nonpolar, aromatic, polar, positively charged, and negatively charged categories.
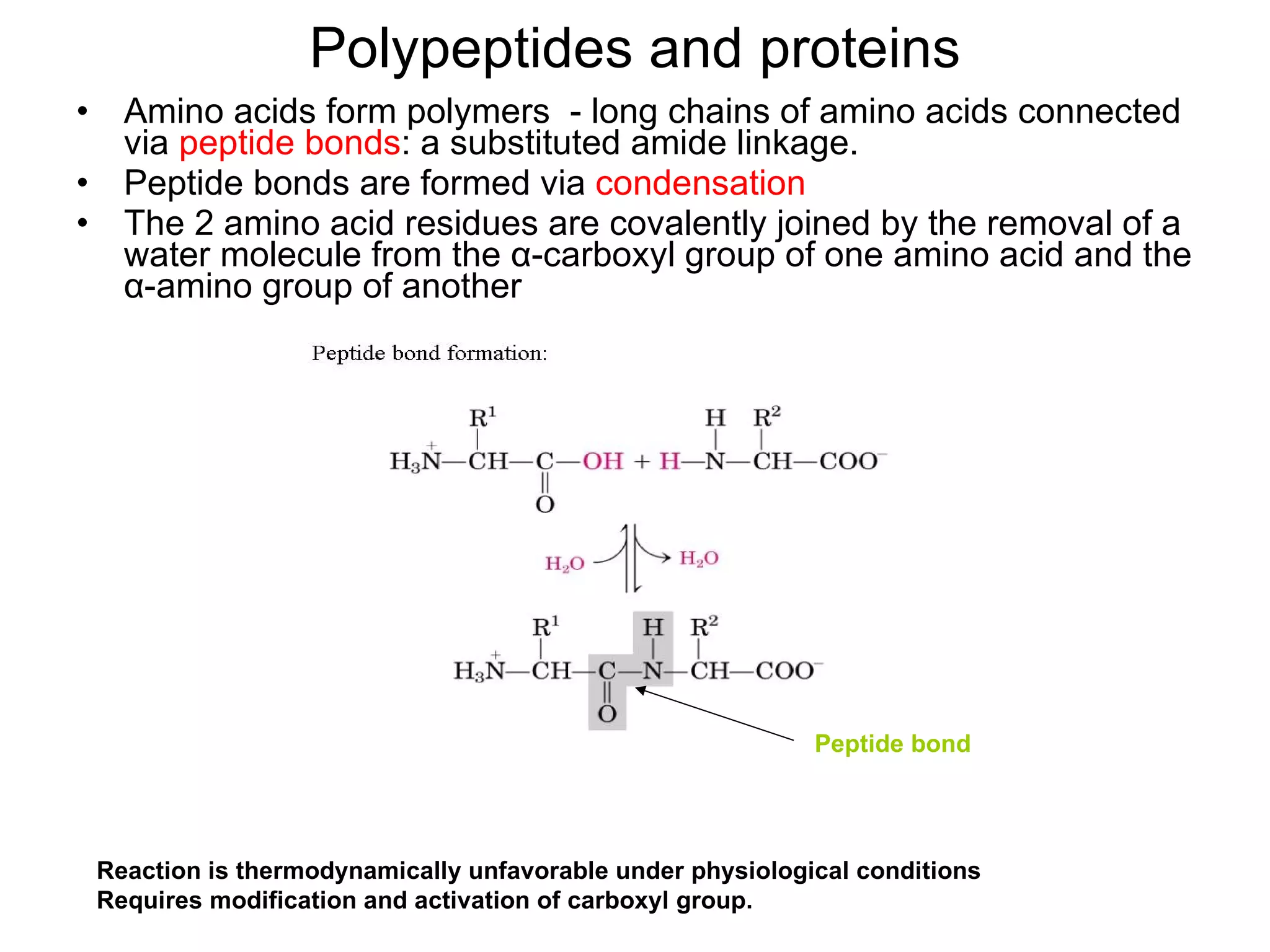
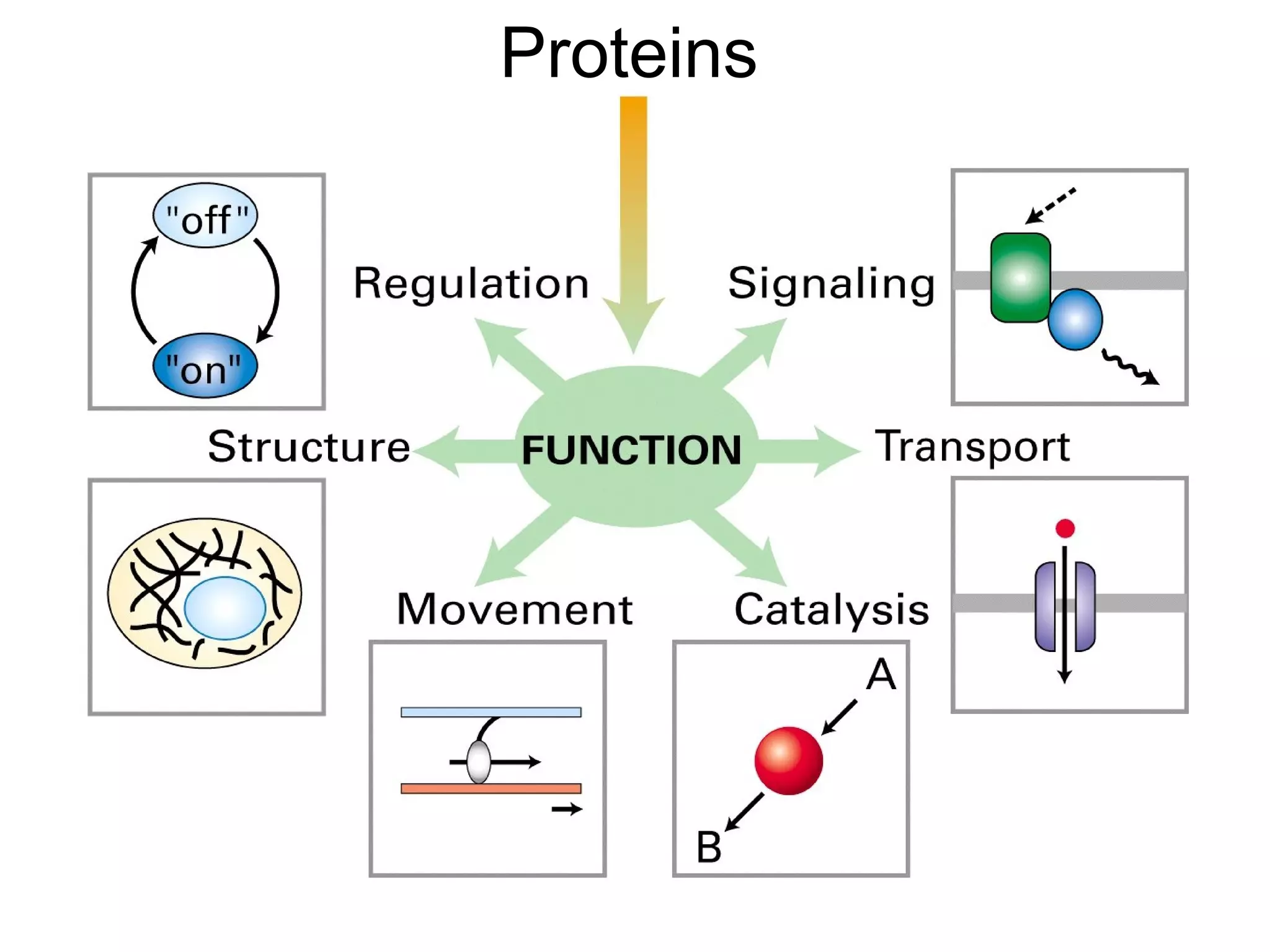
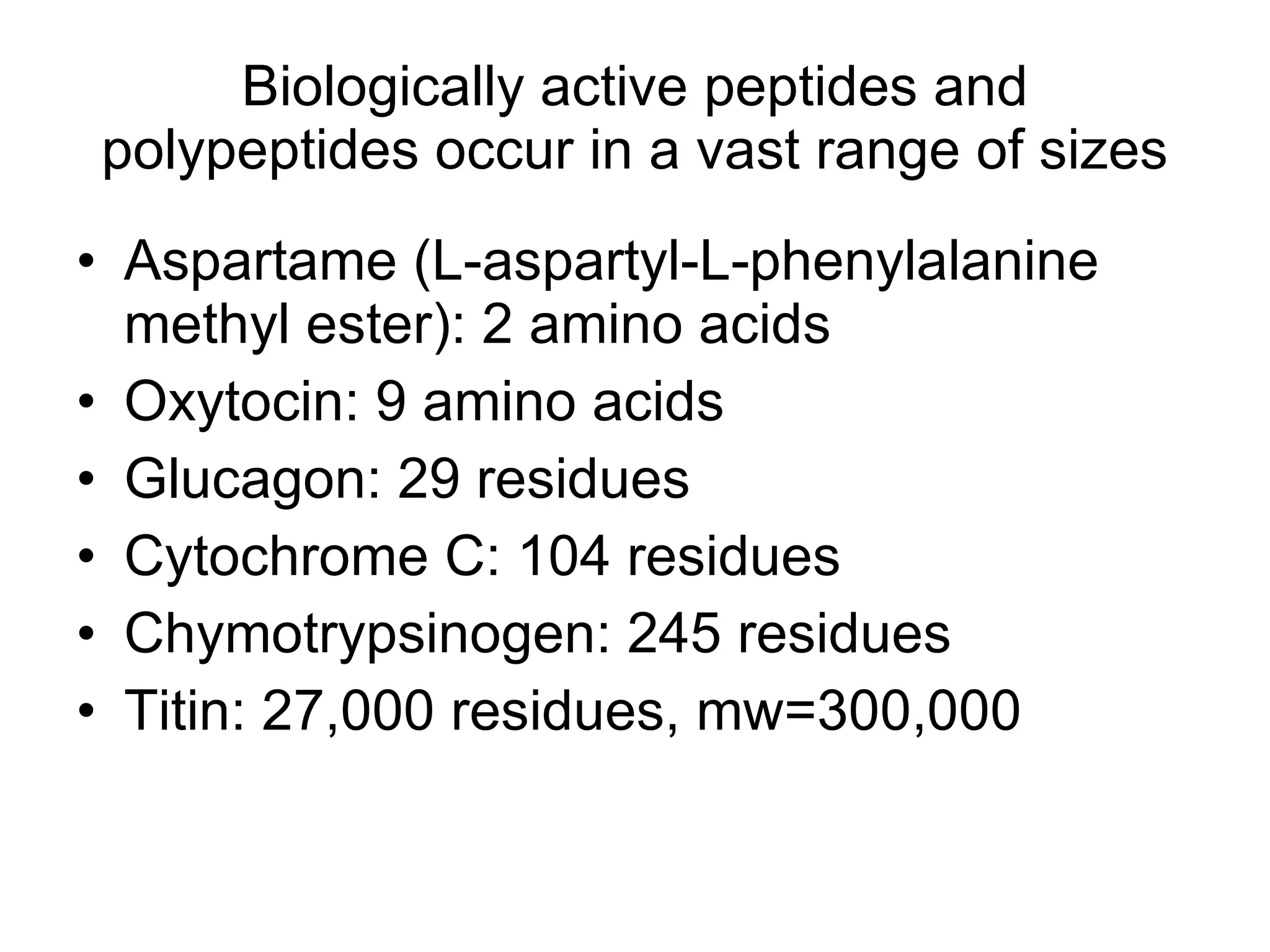
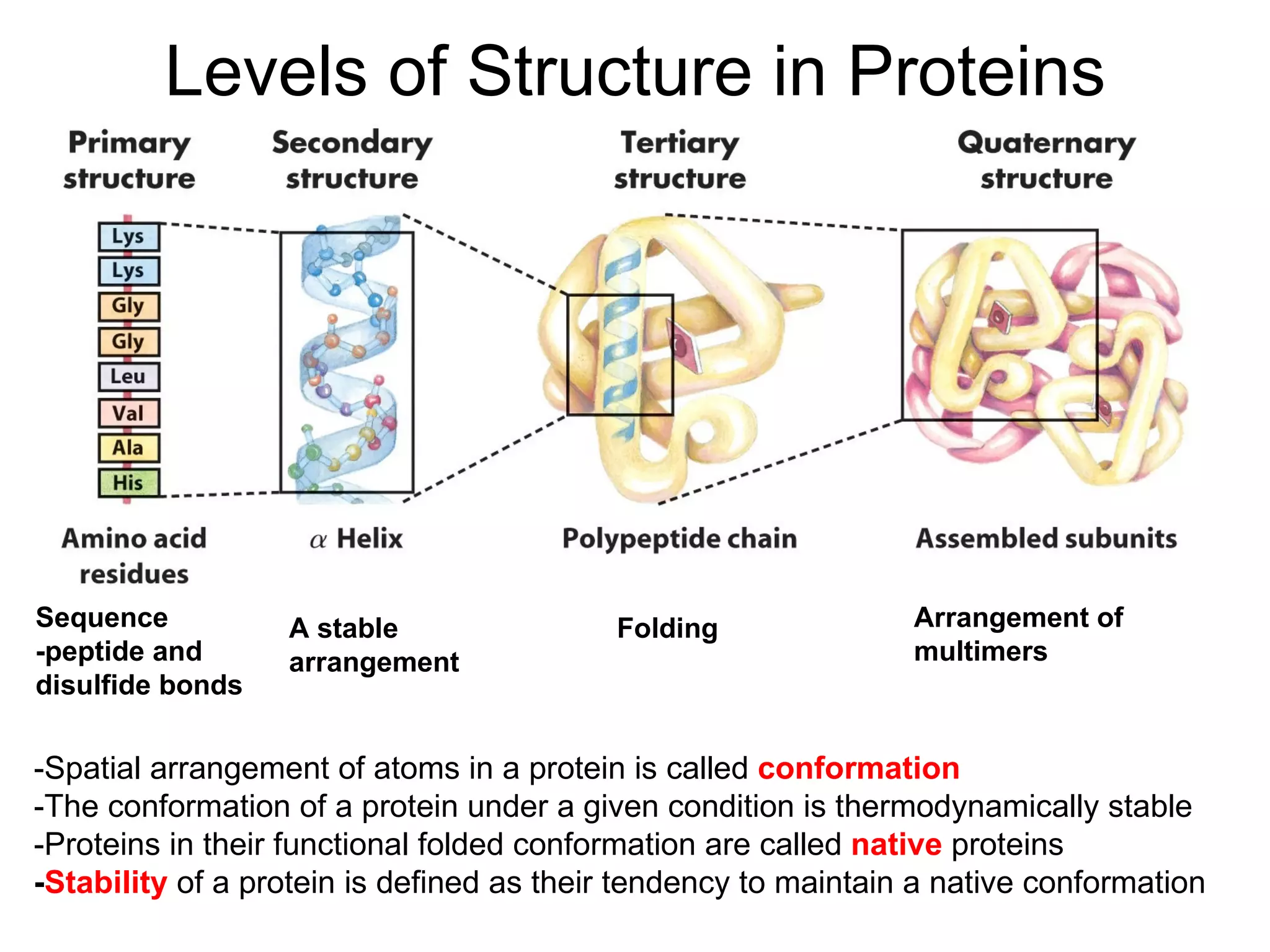
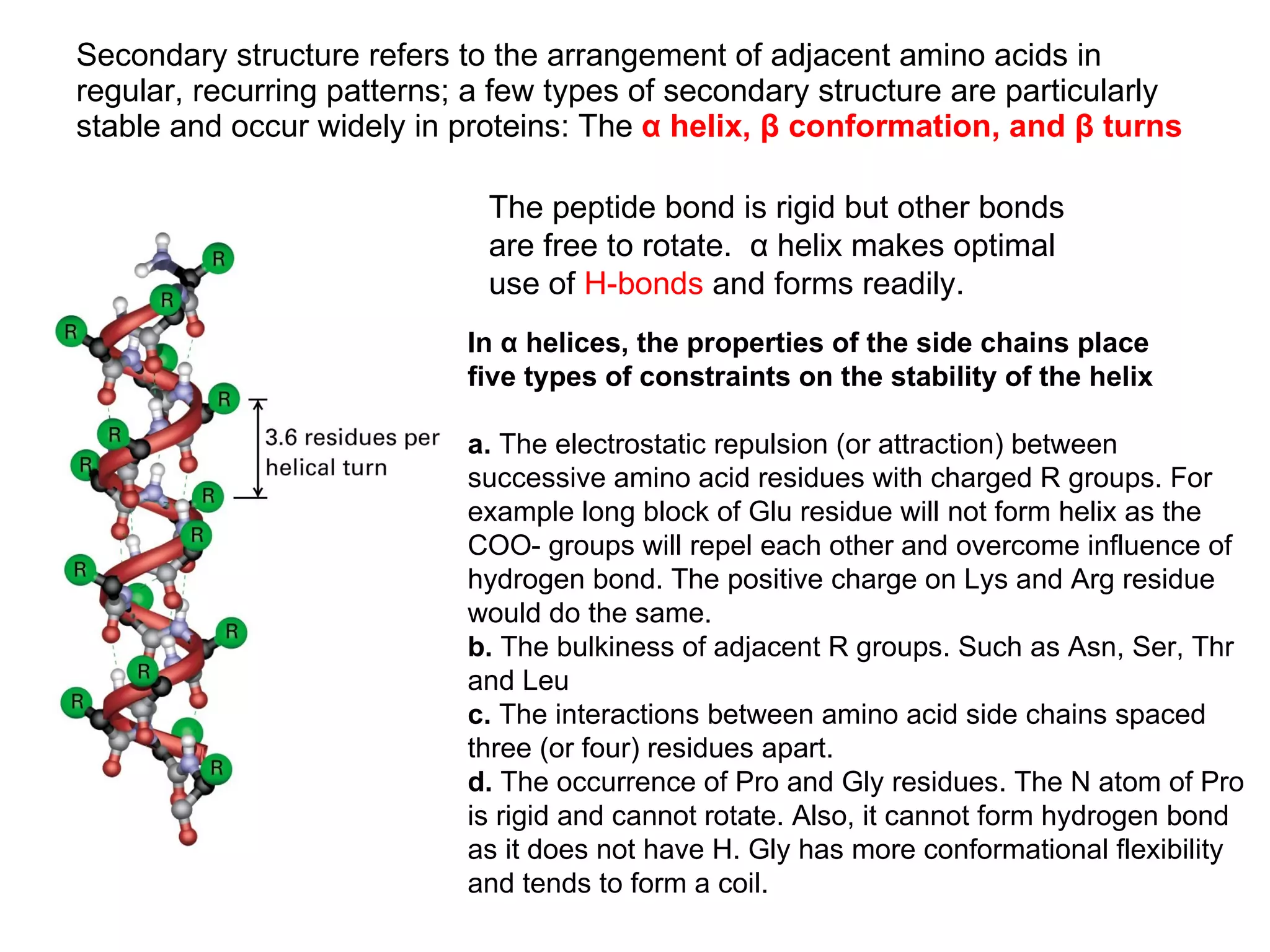
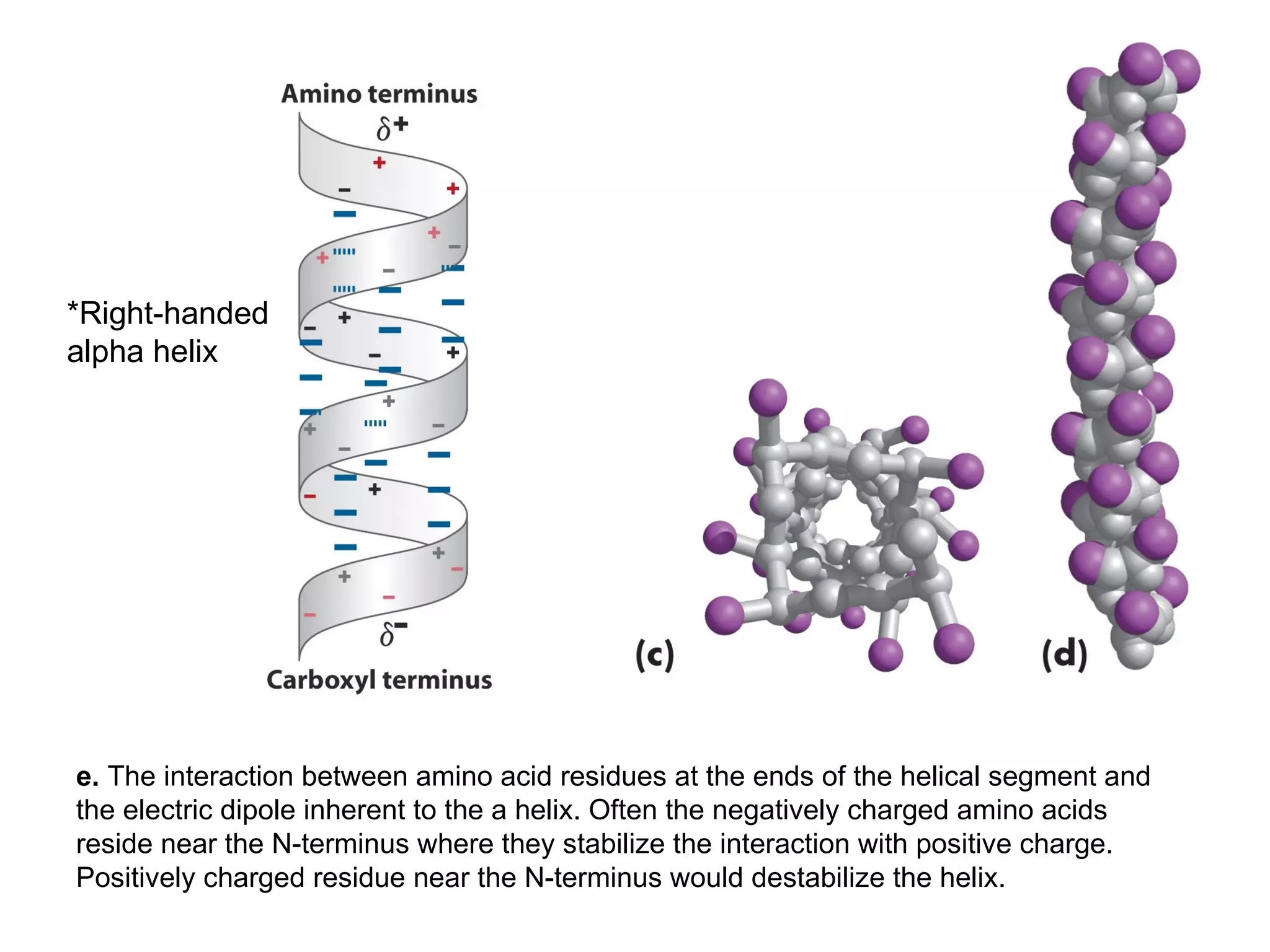
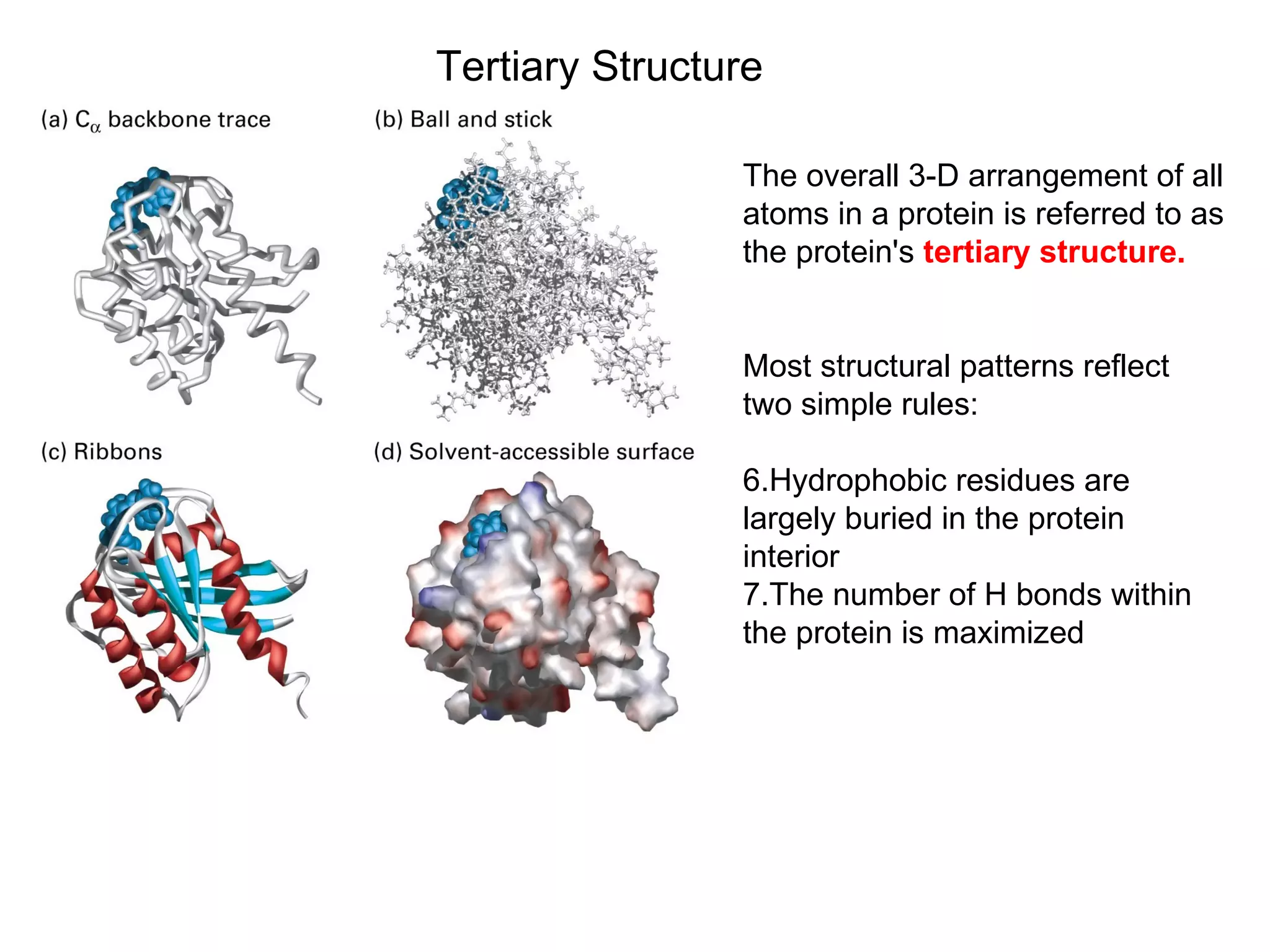
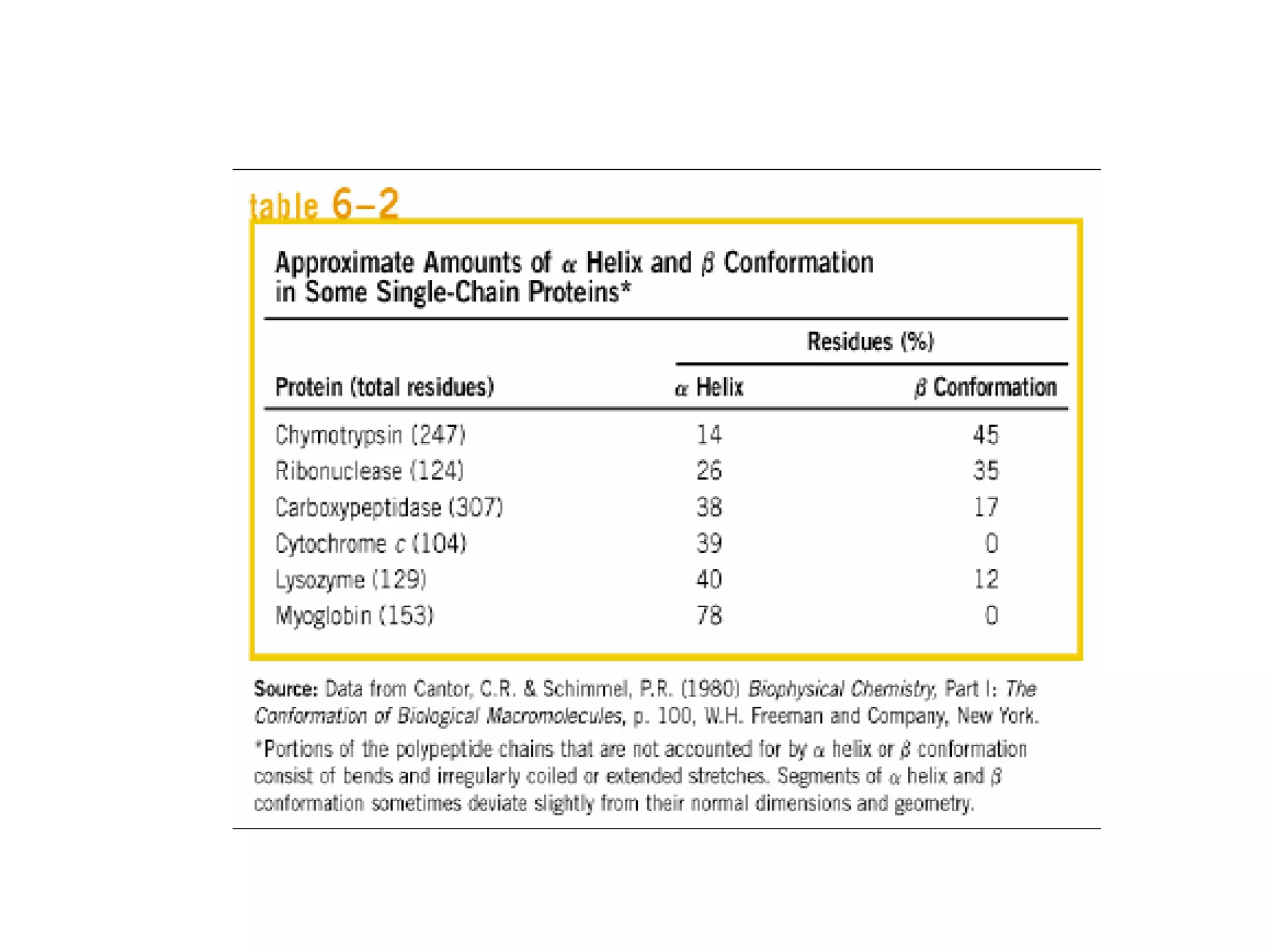
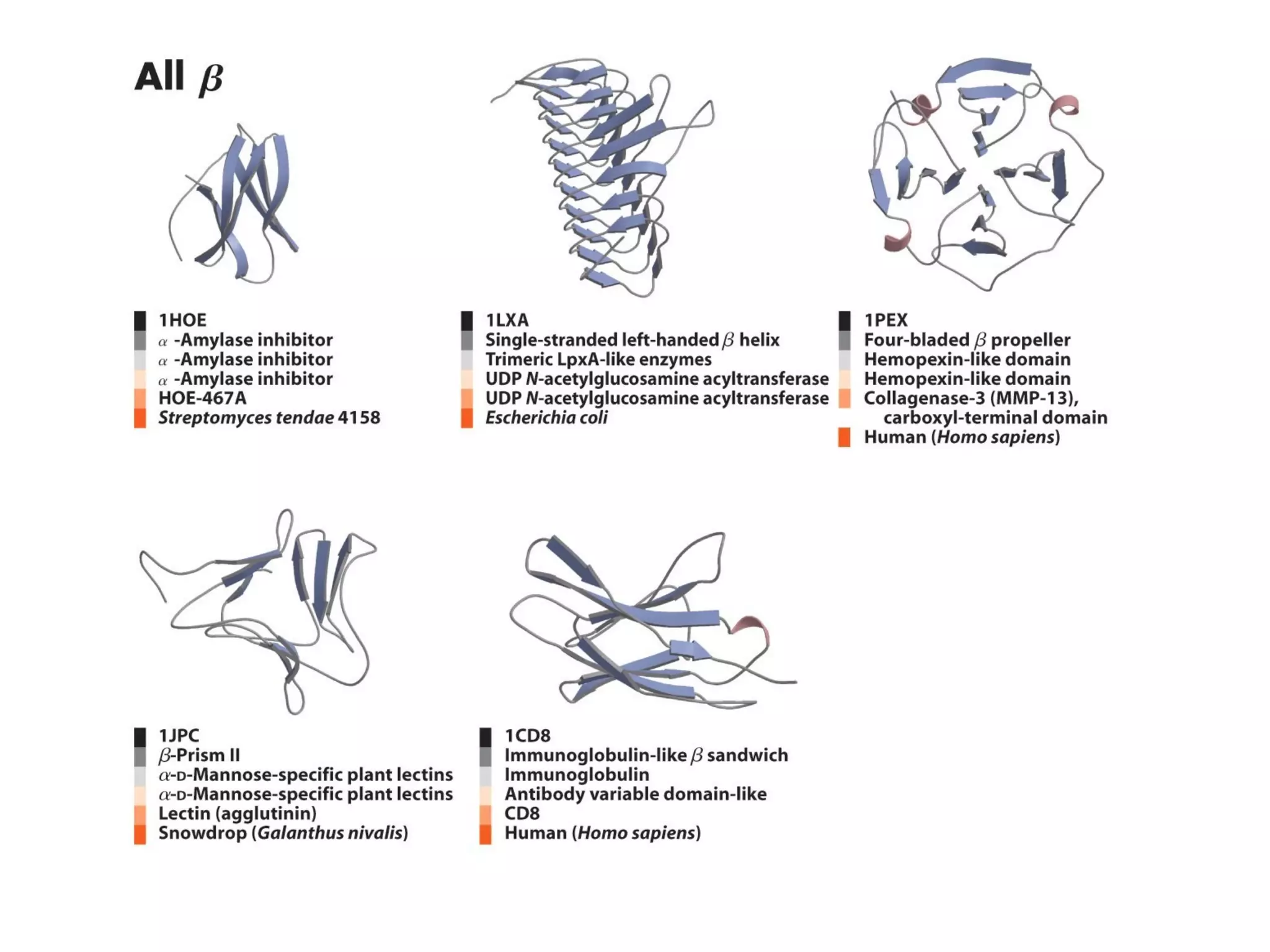
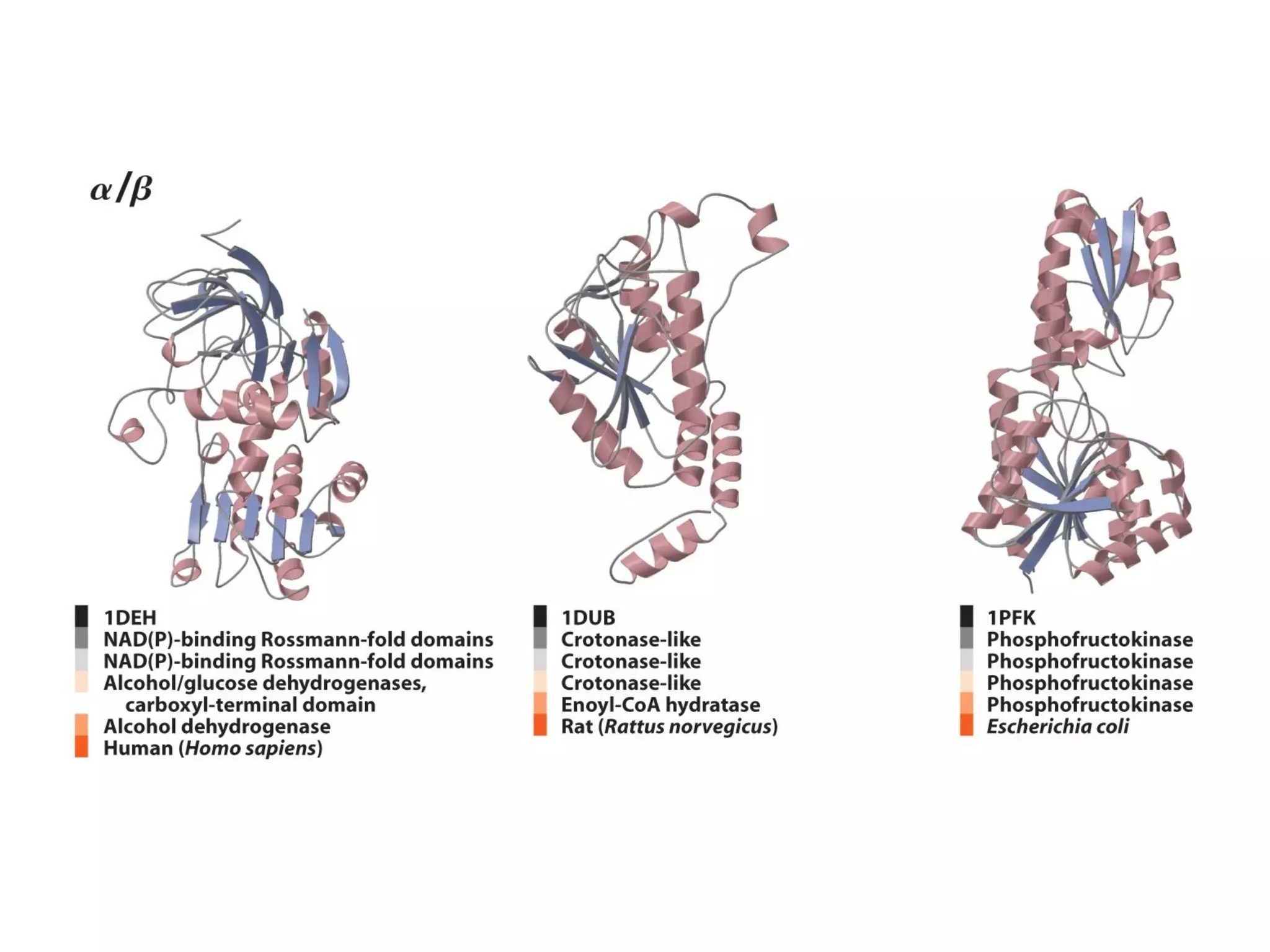
Protein structure is hierarchical, progressing from primary to secondary, tertiary, and quaternary levels. The primary structure is the amino acid sequence. Secondary structures include alpha helices, beta sheets, and turns formed by hydrogen bonding. Tertiary structure refers to the overall 3