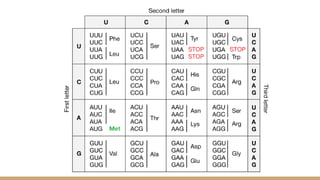
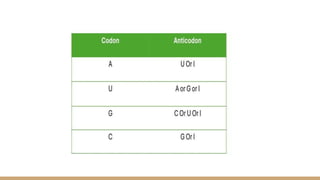
The genetic code consists of codons that translate mRNA sequences into amino acids for protein synthesis, characterized by properties such as degeneracy, universality, and specificity. The wobble hypothesis explains how a limited number of tRNA molecules can recognize multiple codons for the same amino acid, facilitating efficient translation and reducing translational errors. This flexibility contributes to genetic robustness, evolutionary adaptability, and optimized cellular resource management.