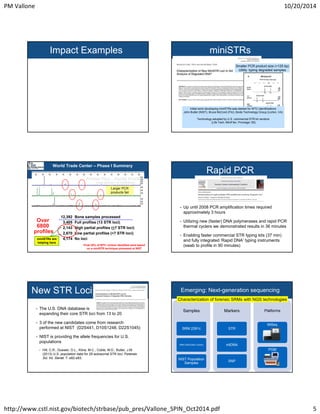
The document outlines a workshop led by Peter M. Vallone on human identity testing within the applied genetics group, emphasizing forensic DNA analysis through various techniques such as PCR, genotyping, and next-generation sequencing. It discusses the applications of human identity testing, the significance of standard reference materials for quality assurance in laboratories, and recent technological advancements in DNA profiling methods. Additionally, it highlights the role of interlaboratory studies in improving reproducibility across different labs.


![PM Vallone 10/20/2014
Purpose of an Interlaboratory Study
Interlaboratory studies (ILS) are a way
for multiple laboratories to compare
results and demonstrate that the
methods or instrument platforms used
in one's own laboratory are
reproducible in another laboratory
NIST Initiated Interlaboratory Studies
ILS Participating
Labs Publications/Dissemination
Evaluation of CSF1PO,
TPOX, and TH01 34
Kline MC, Duewer DL, Newall P, Redman JW, Reeder DJ, Richard M.
(1997) Interlaboratory evaluation of STR triplex CTT. J. Forensic Sci. 42:
897-906
Mixed Stain Studies
#1 and #2 (1997 and 1999)
45
Duewer DL, Kline MC, Redman JW, Newall PJ, Reeder DJ. (2001) NIST
Mixed Stain Studies #1 and #2: interlaboratory comparison of DNA
quantification practice and short tandem repeat multiplex performance with
multiple-source samples. J. Forensic Sci. 46: 1199-1210
The ILS lead to publications, reference materials,
and dissemination of results to the community
Independent measure of how the DNA typing
Mixed Stain Study #3
(2000 and 2001) 74
Kline, M.C., Duewer, D.L., Redman, J.W., Butler, J.M. (2003) NIST mixed
stain study 3: DNA quantitation accuracy and its influence on short tandem
repeat multiplex signal intensity. Anal. Chem. 75: 2463-2469.
Duewer, D.L., Kline, M.C., Redman, J.W., Butler, J.M. (2004) NIST Mixed
Stain Study #3: signal intensity balance in commercial short tandem repeat
multiplexes, Anal. Chem. 76: 6928-6934.
DNA Quantitation Study
80 Kline, M.C., Duewer, D.L., Redman, J.W., Butler, J.M. (2005) Results from
(2004) the NIST 2004 DNA Quantitation Study, J. Forensic Sci. 50(3):571-578
Mixture Interpretation
Study (2005)
community is performing
69 http://www.cstl.nist.gov/strbase/interlab/MIX05/MIX05poster.pdf
Mixture Interpretation
Study (2013) 108 Manuscript in preparation
Rapid DNA Testing
(Fall 2013) 3 http://www.cstl.nist.gov/strbase/pub_pres/Vallone_BCC_Talk_Sept2013.pdf
Individual Performance in an Interlaboratory
Study
DNA Quantitation
10
5
3
1
0.5
0.3
R S T V X U W
Various samples (R-W)
97.5%
75%
Median
25%
2.5%
DNA Concentration, ng / L
Overall results
Individual lab performance
Modified plot from Kline, M.C., et al. (2003) Anal. Chem. 75: 2463-2469
NIST Mixed Stain Study 3: DNA Quantitation Accuracy and Its Influence on Short Tandem Repeat
Multiplex Signal Intensity
Results…
• Quantitation Methods and Frequency of Use
• Interpretation of Semi-Quantitative Data
• Method Sensitivities
• Combining Within-Analyst Replicates and Within-laboratory Duplicates
• Distributions of the Among-laboratory Results
• Measurement Variability
• Measurement Performance Characteristics
• Consensus Values and Variability as Functions of [DNA]
• Blot-Based Vs. Q-PCR Methods
• Single-Source Vs. Multiple-Source Materials
• Polypropylene Vs. Teflon Sample Containers
Education and Training
http://www.cstl.nist.gov/strbase/
Since 10/1997
Information related to:
Basics of STR typing
Variant allele reports
STR typing kit
STR loci ‘fact sheets’
Population data
Software tools
Team outputs:
Since 1998 ≈ 200 papers
Since 2000 ≈ 865 posters/
presentations/workshops
Textbooks Written by Dr. John Butler
http://www.cstl.nist.gov/biotech/strbase/pub_pres/Vallone_SPIN_Oct2014.pdf 4](https://image.slidesharecdn.com/vallonespinoct2014-141209161936-conversion-gate02/85/Forensics-Human-Identity-Testing-in-the-Applied-Genetics-Group-4-320.jpg)