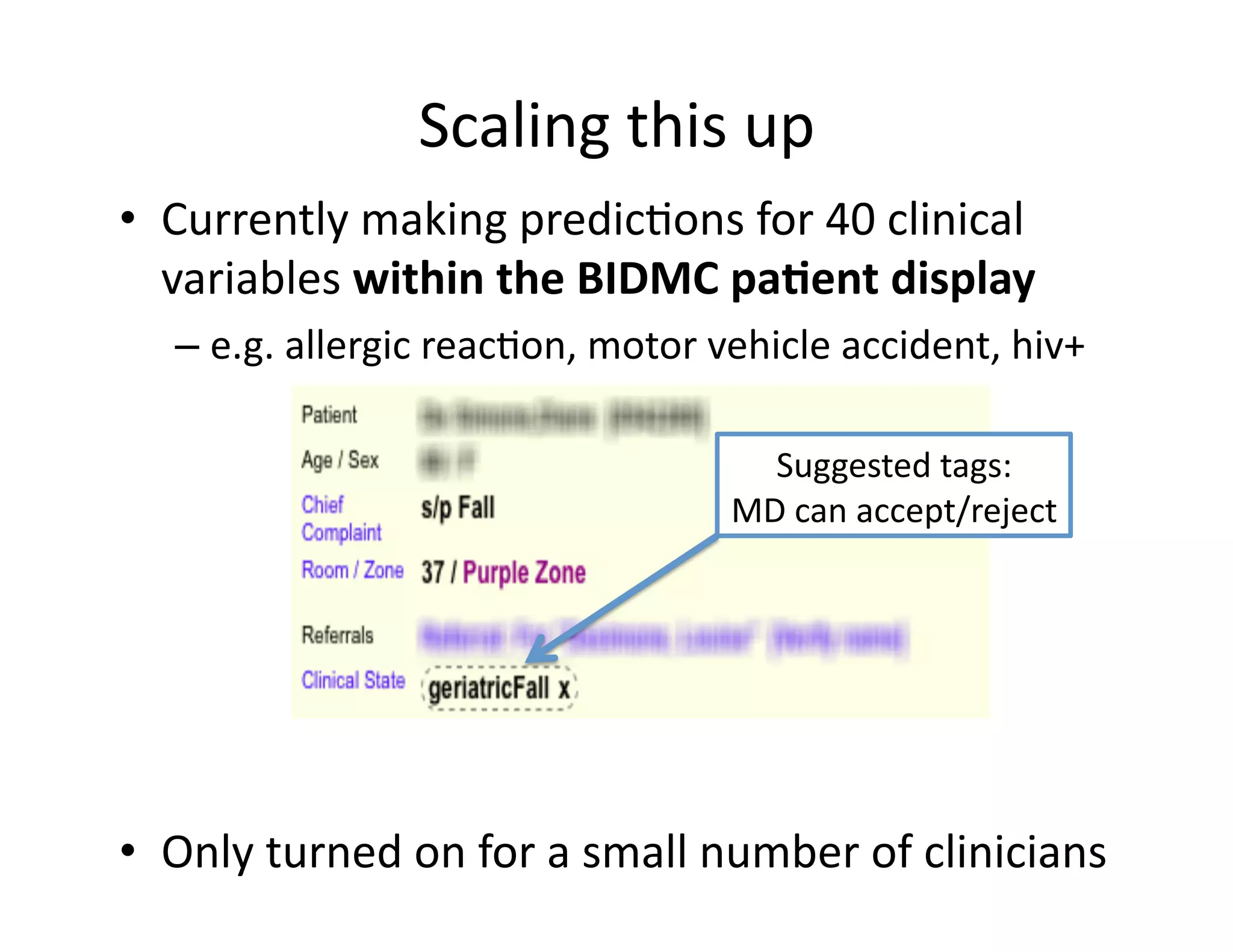
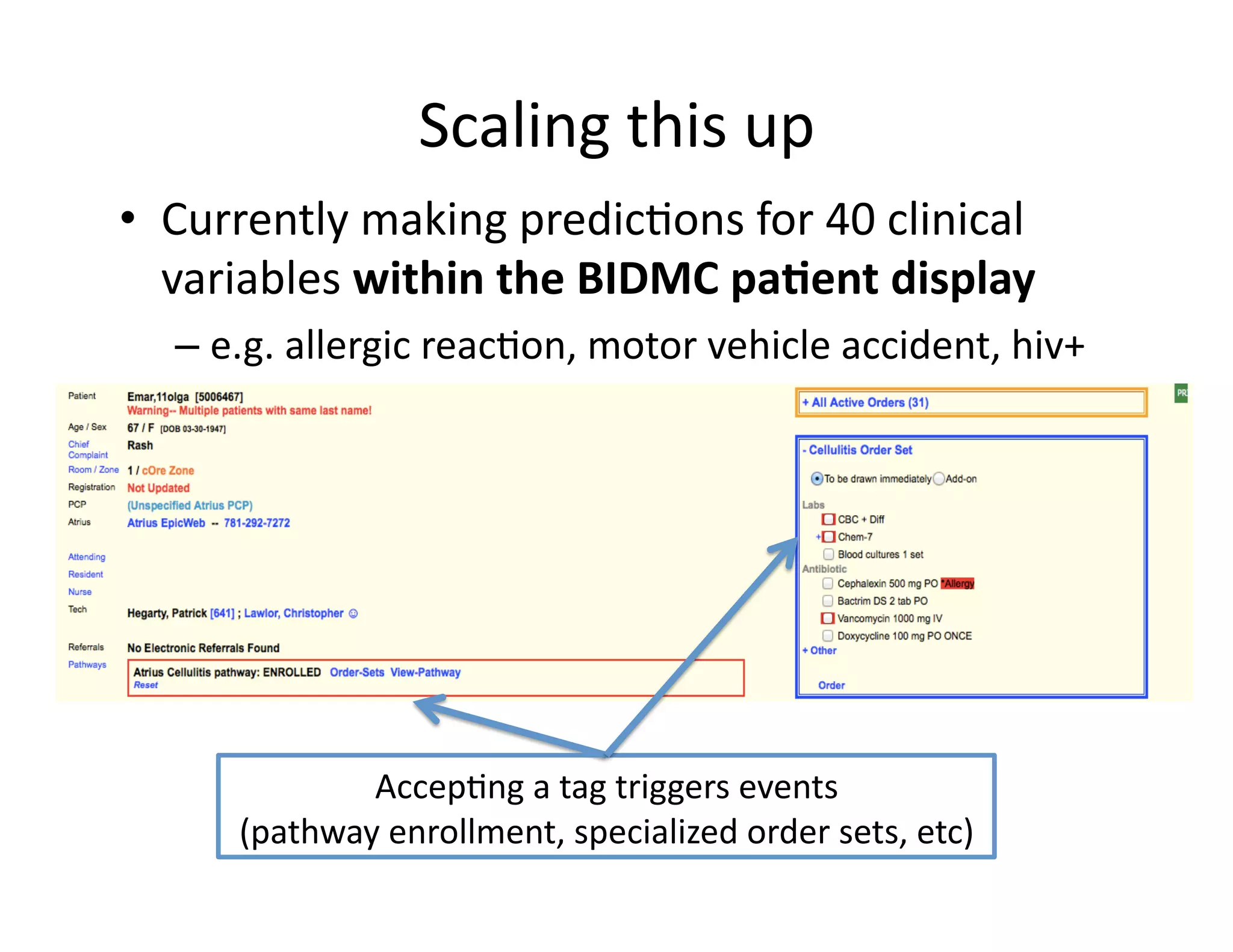
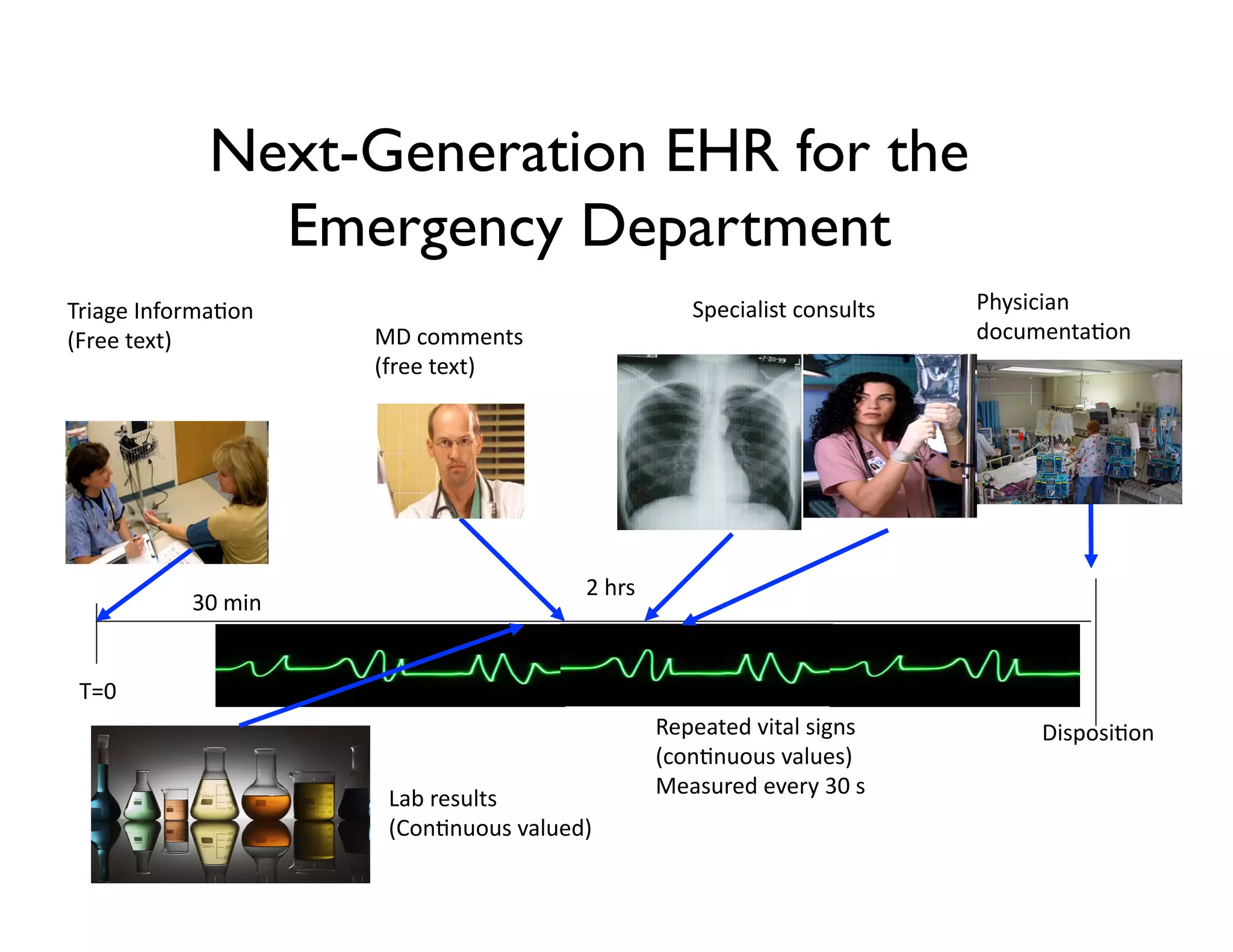
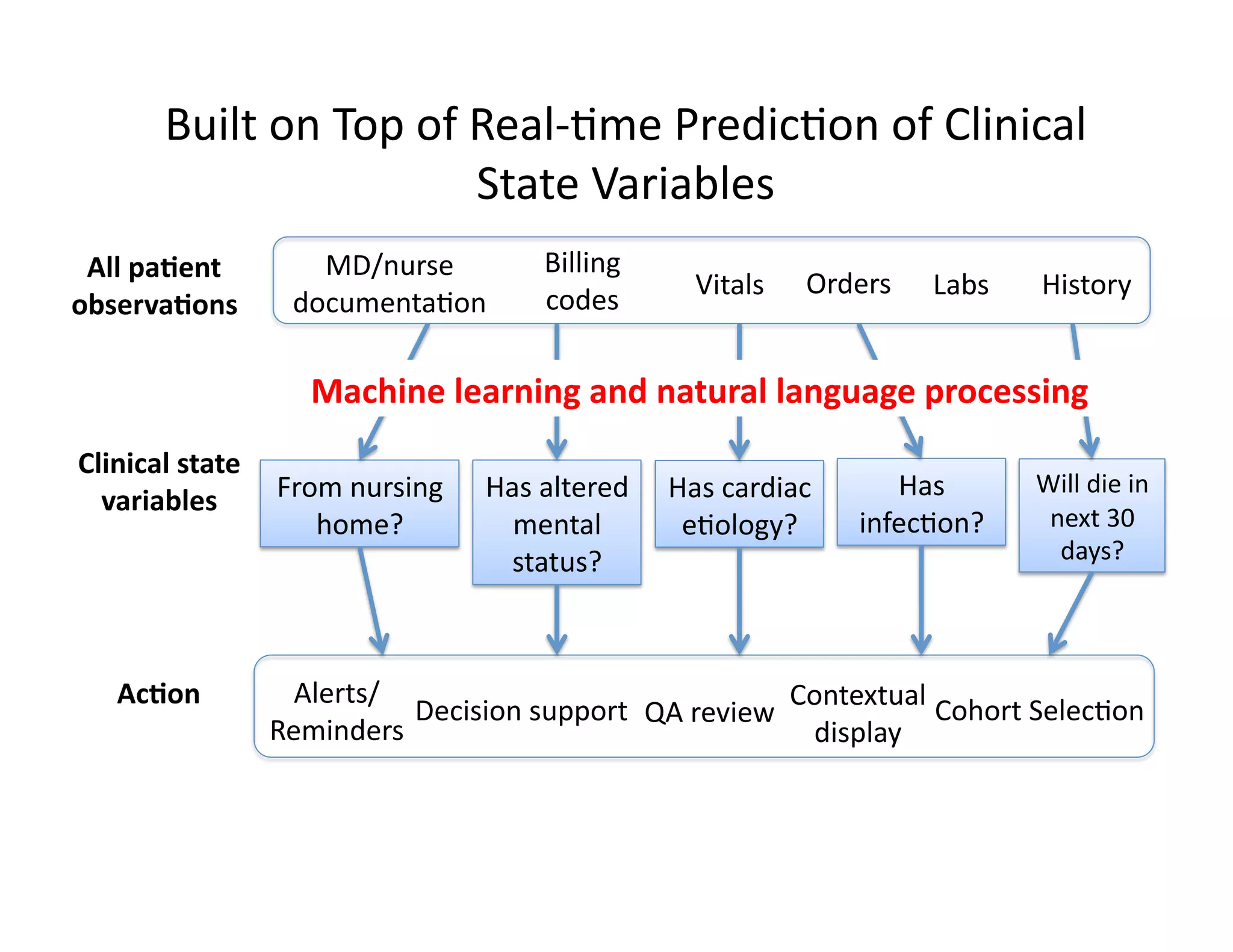
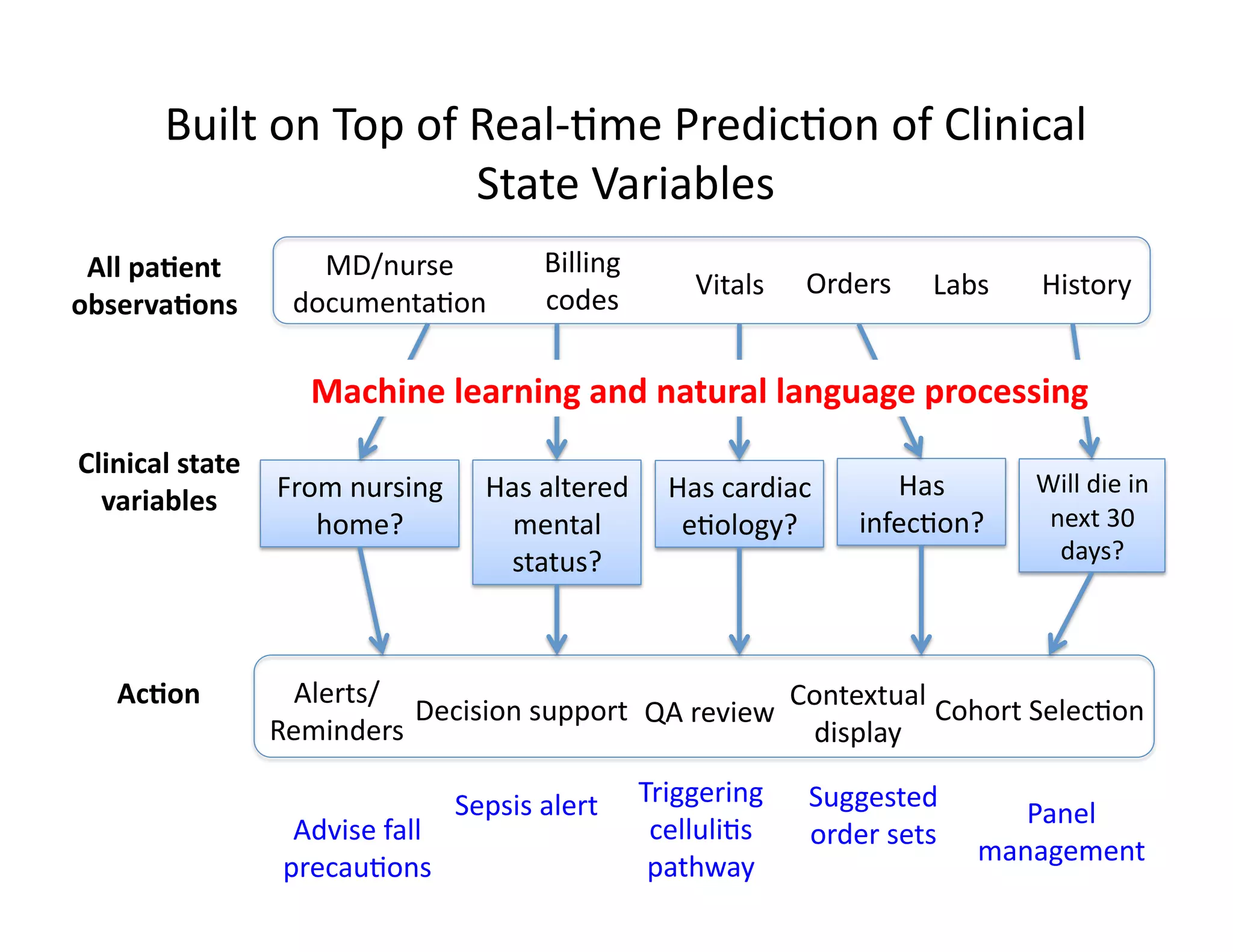
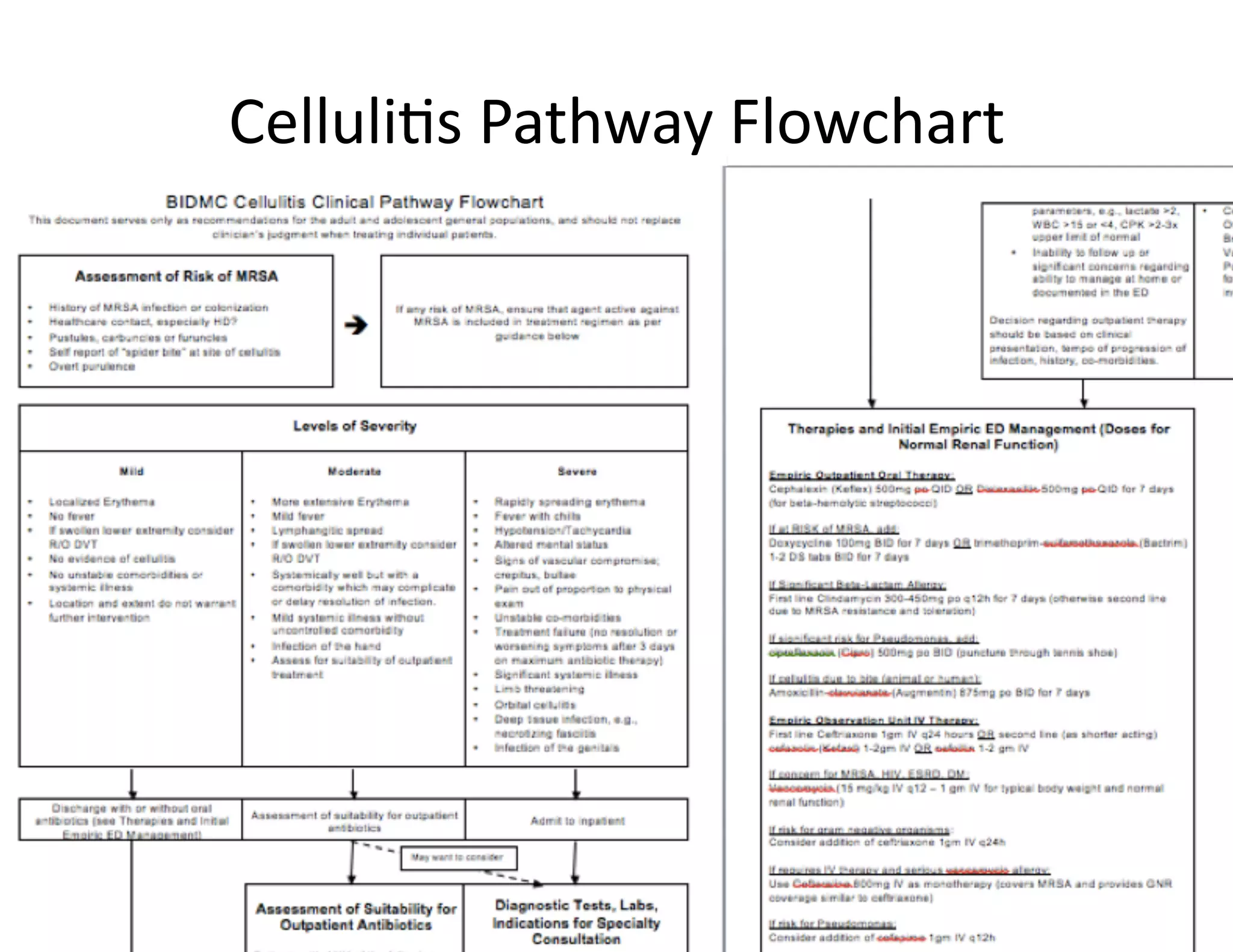
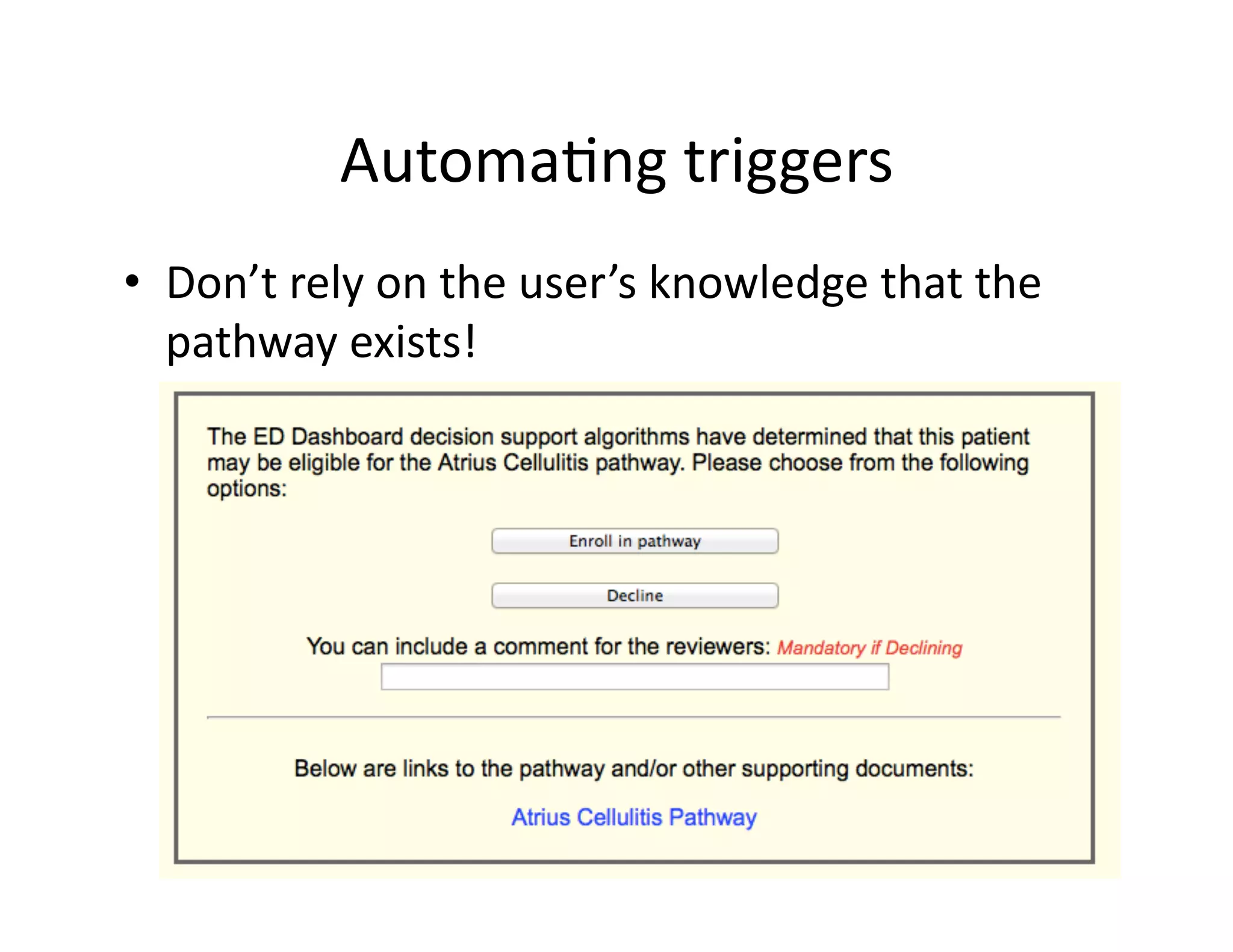
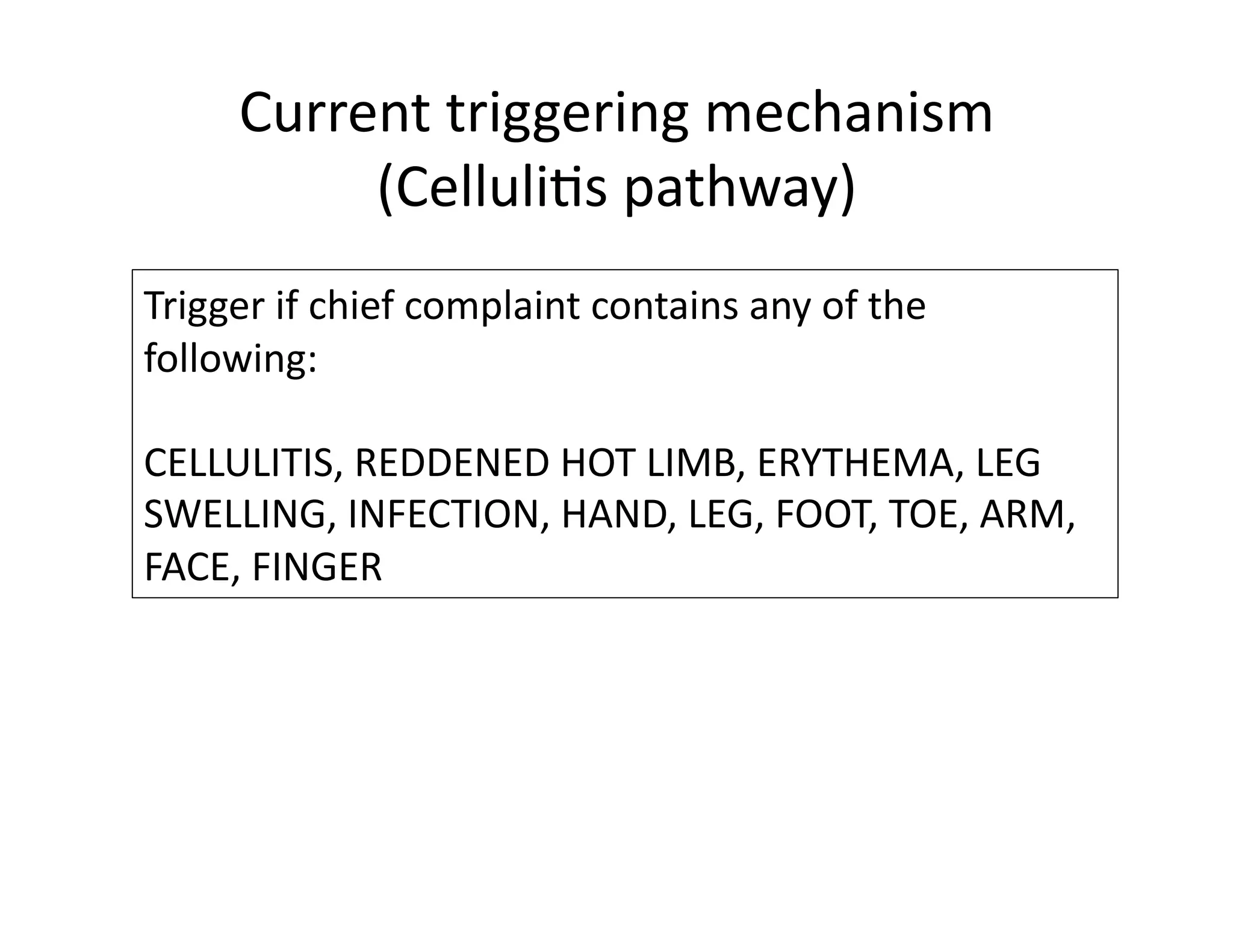
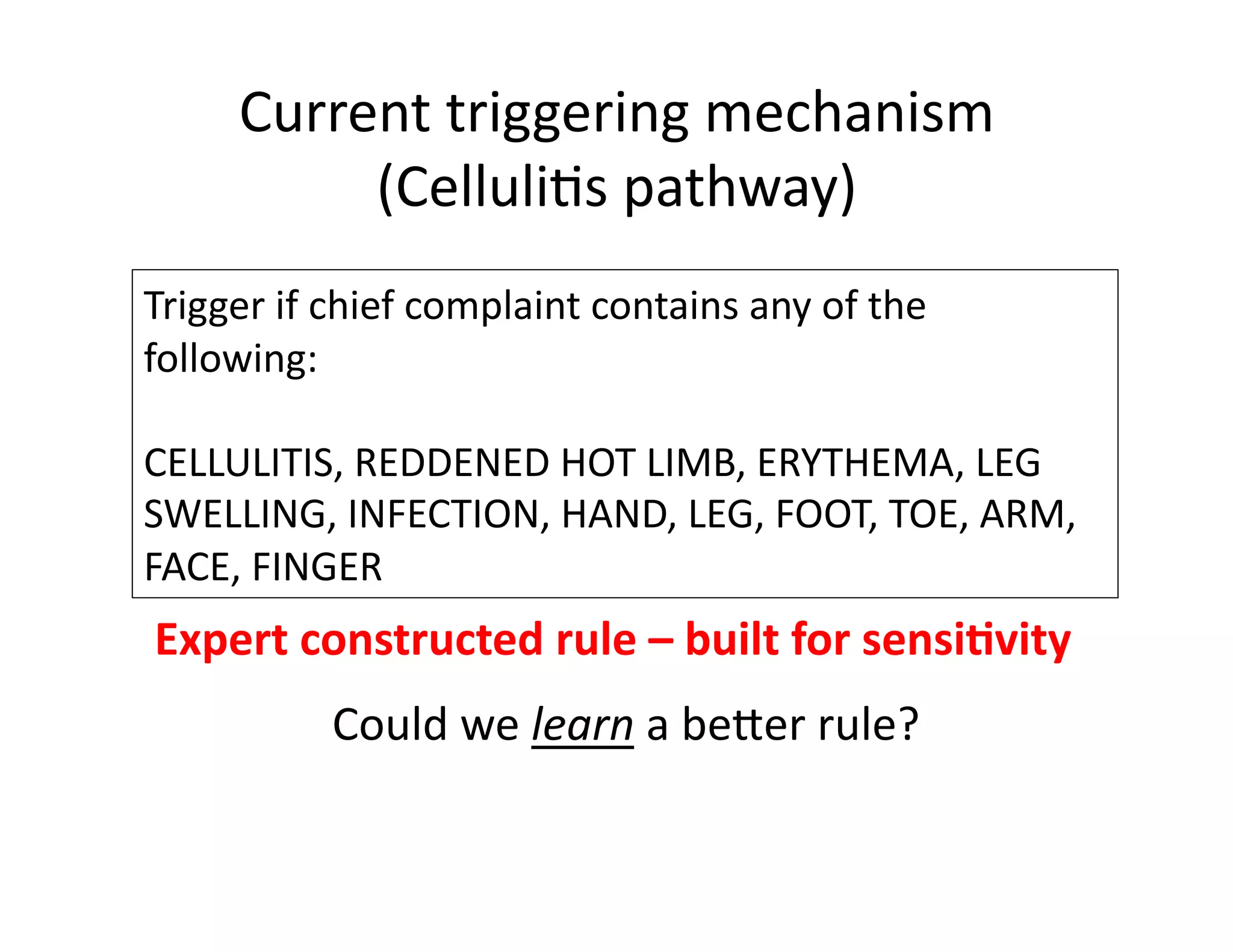
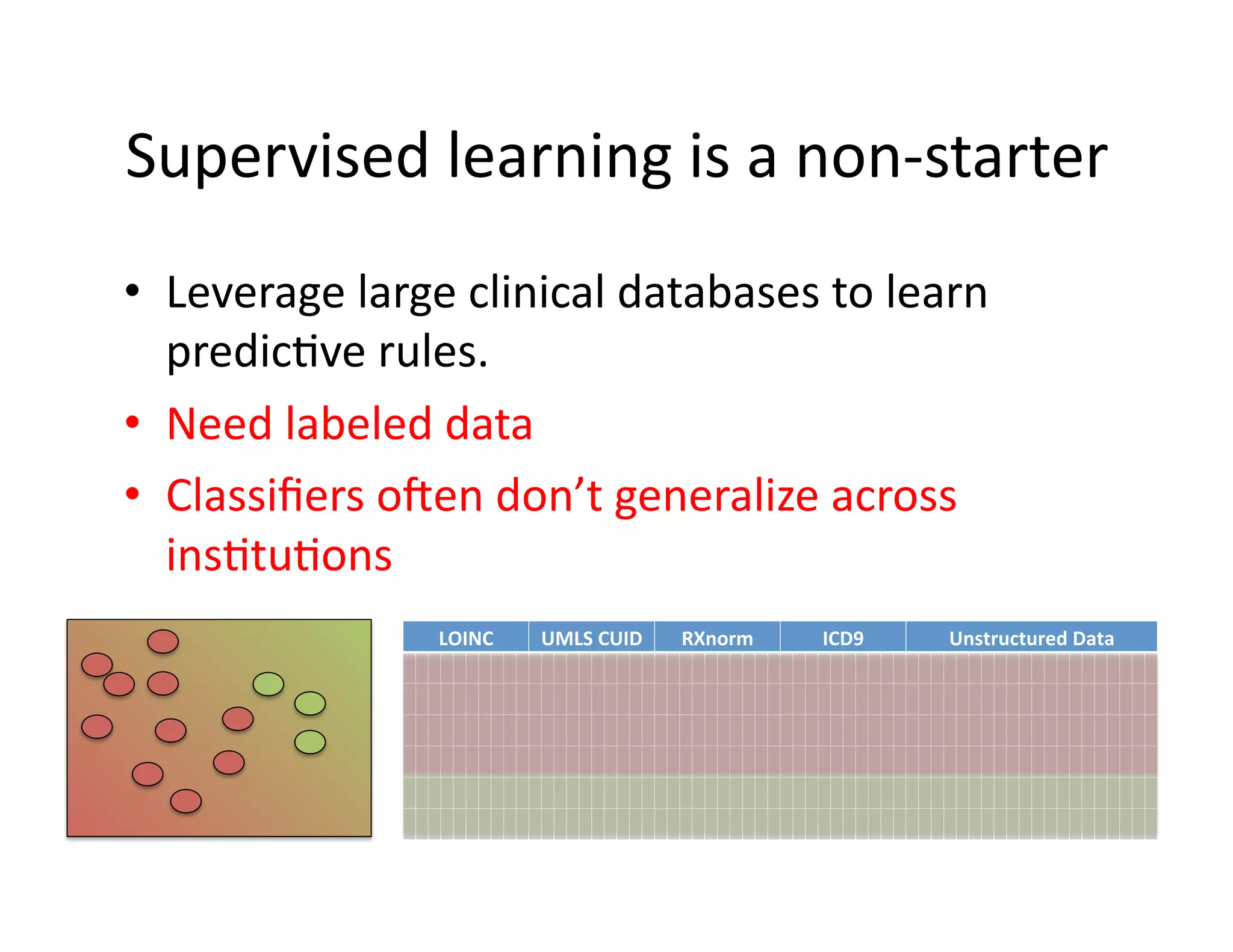
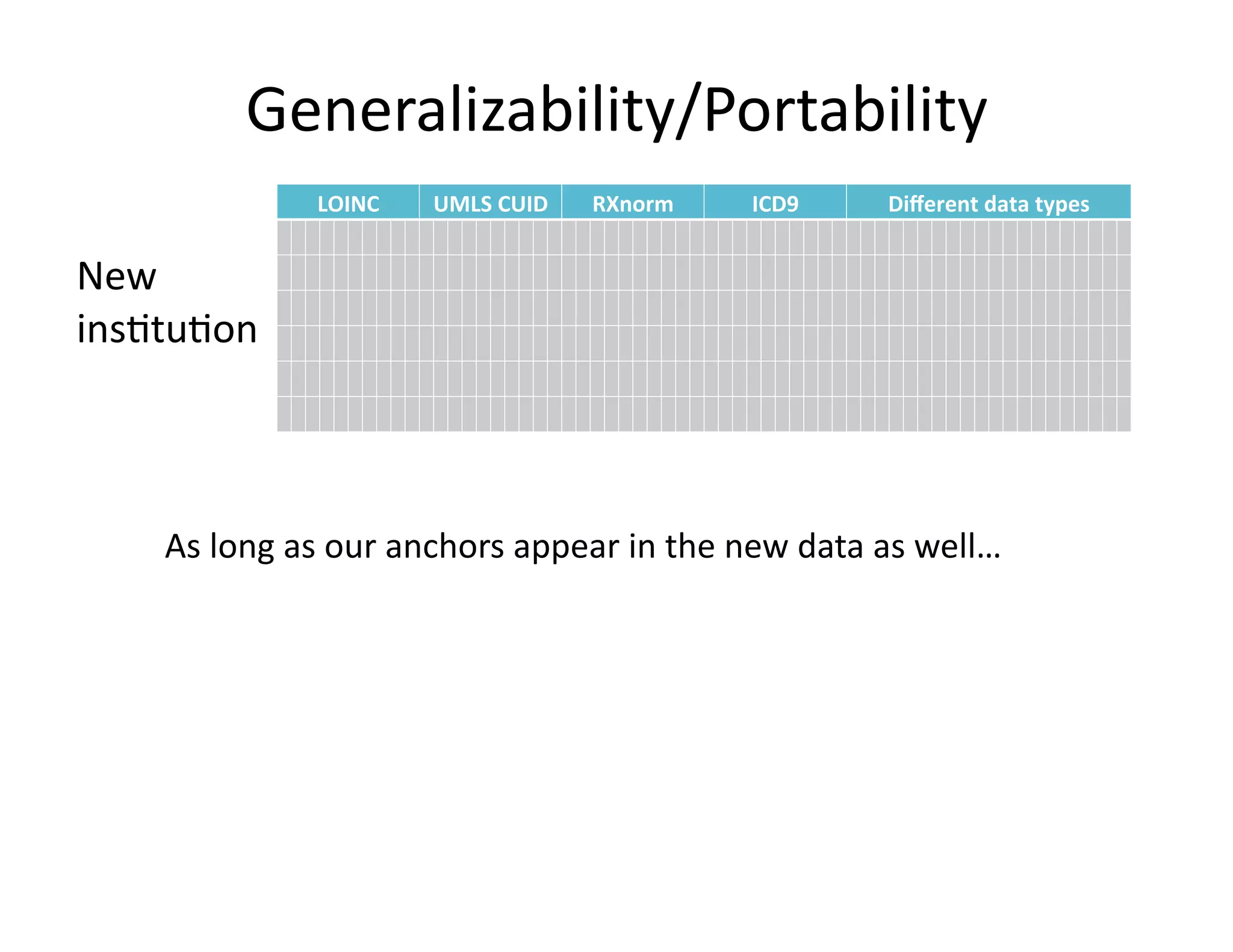
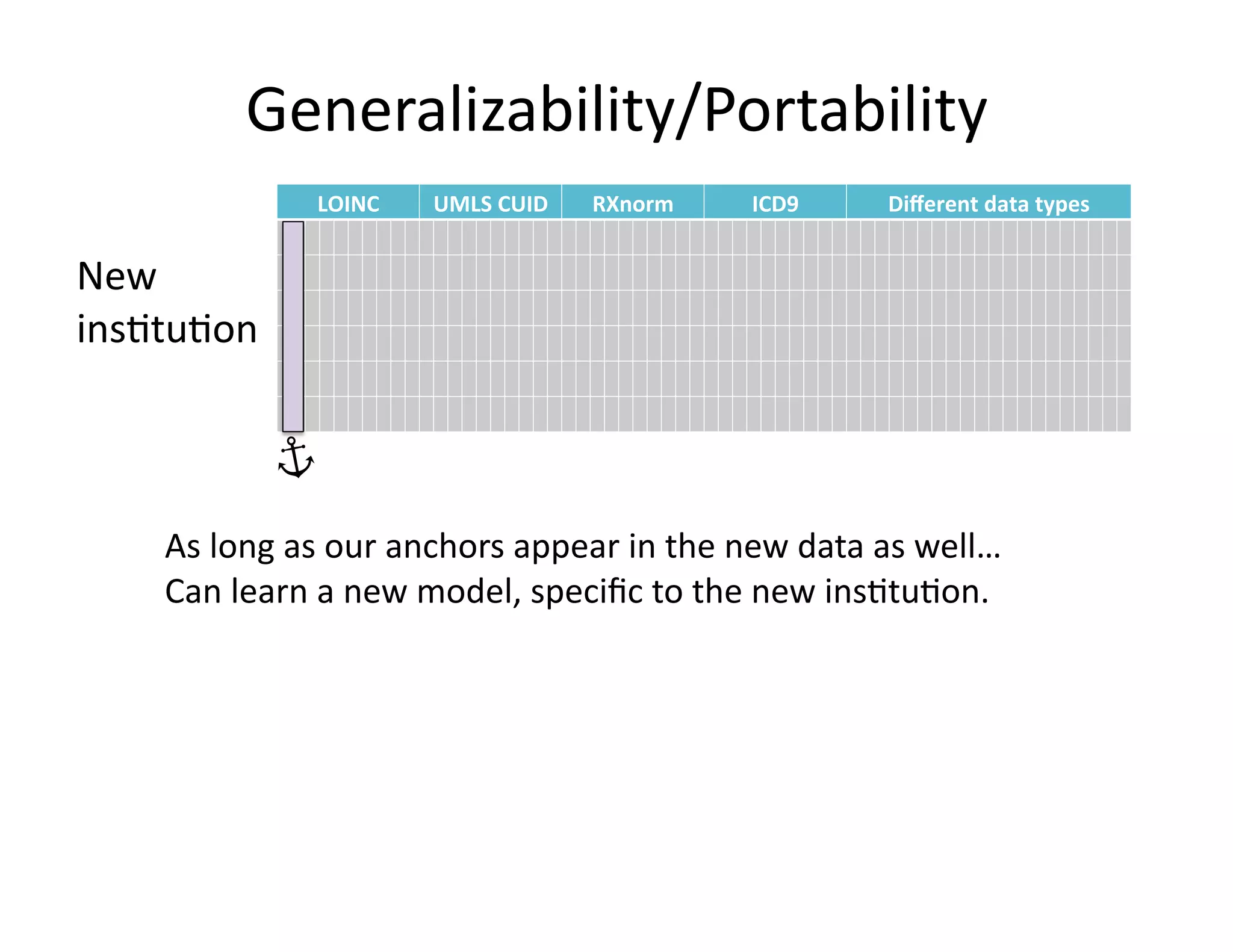
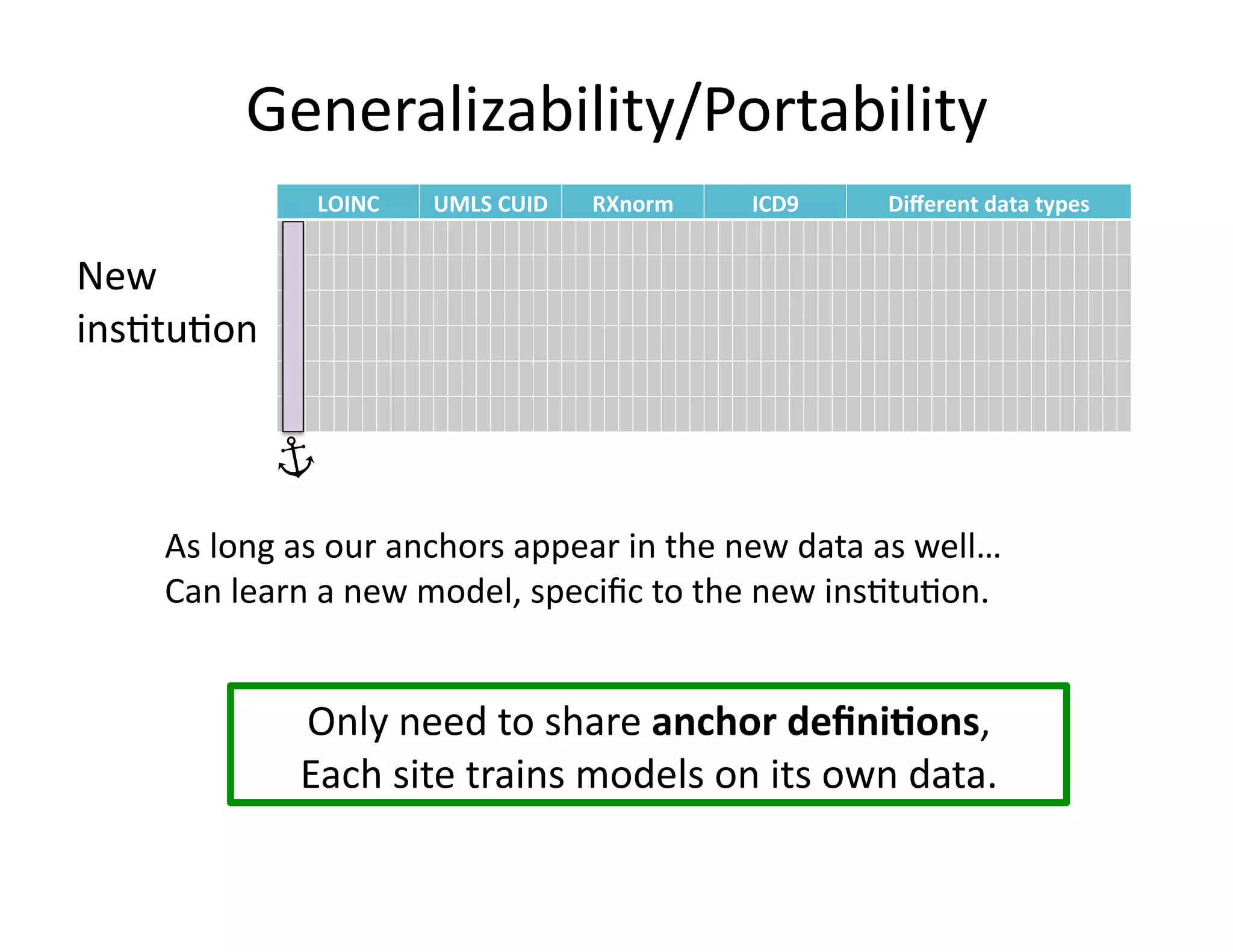
The document discusses using machine learning to automate clinical pathways in emergency departments. It describes developing a next-generation electronic health record system that predicts clinical state variables in real-time from patient data to provide alerts, decision support, and trigger clinical pathways. Specifically, it discusses using an "anchor and learn" framework to improve on an existing rule for triggering a cellulitis pathway, by combining simple expert rules with analysis of large clinical databases that does not require manual labeling of data.
![Health
Informa@on
Technology
is
Rapidly
Changing
• Aided
by
HITECH
Act,
hospital
adop@on
of
EHRs
has
increased
5-‐fold
since
2008
[Charles
et
al.,
ONC
Data
Brief,
May
2014]](https://image.slidesharecdn.com/ibmcognitivecomputingsept162015-150917131628-lva1-app6892/75/Using-Machine-Learning-to-Automate-Clinical-Pathways-2-2048.jpg)
![• Over
$4
billion
of
investment
in
digital
health
startups
in
2014
Health
Informa@on
Technology
is
Rapidly
Changing
Analy@cs
/
Big
Data
Healthcare
Consumer
Engagement
[Wang
et
al.,
“Digital
health
funding
in
Q1
2015
over
$600M”,
Rock
Health,
April
2015]
EHR
/
Clinical
Workflow
Digital
Diagnos@cs
Popula@on
Health
Management
Digital
Medical
Device](https://image.slidesharecdn.com/ibmcognitivecomputingsept162015-150917131628-lva1-app6892/75/Using-Machine-Learning-to-Automate-Clinical-Pathways-3-2048.jpg)
![[Weber
et
al.
(2014).
Finding
the
Missing
Link
for
Big
Biomedical
Data.
JAMA.]
Wealth
of
digital
health
data
available](https://image.slidesharecdn.com/ibmcognitivecomputingsept162015-150917131628-lva1-app6892/75/Using-Machine-Learning-to-Automate-Clinical-Pathways-4-2048.jpg)


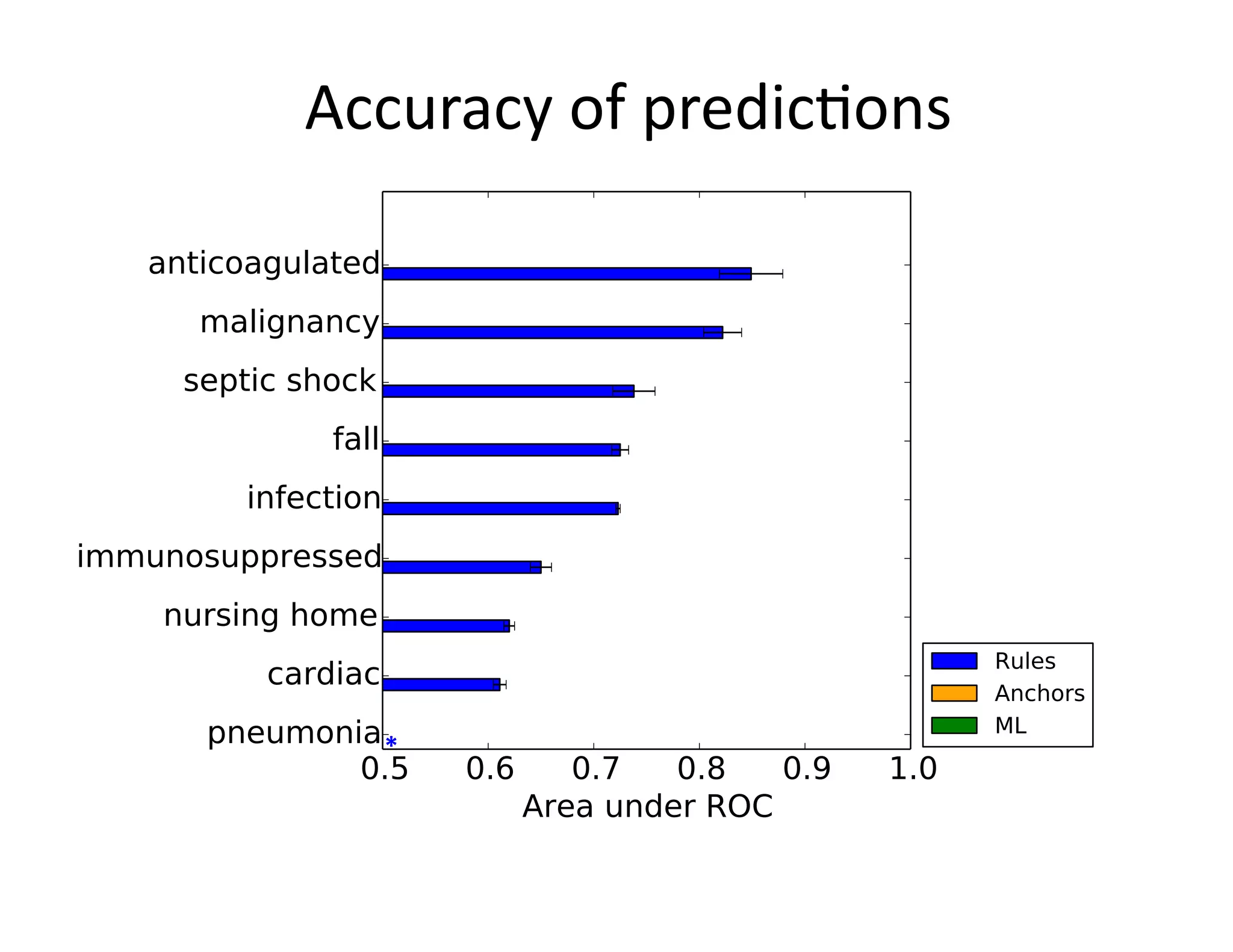
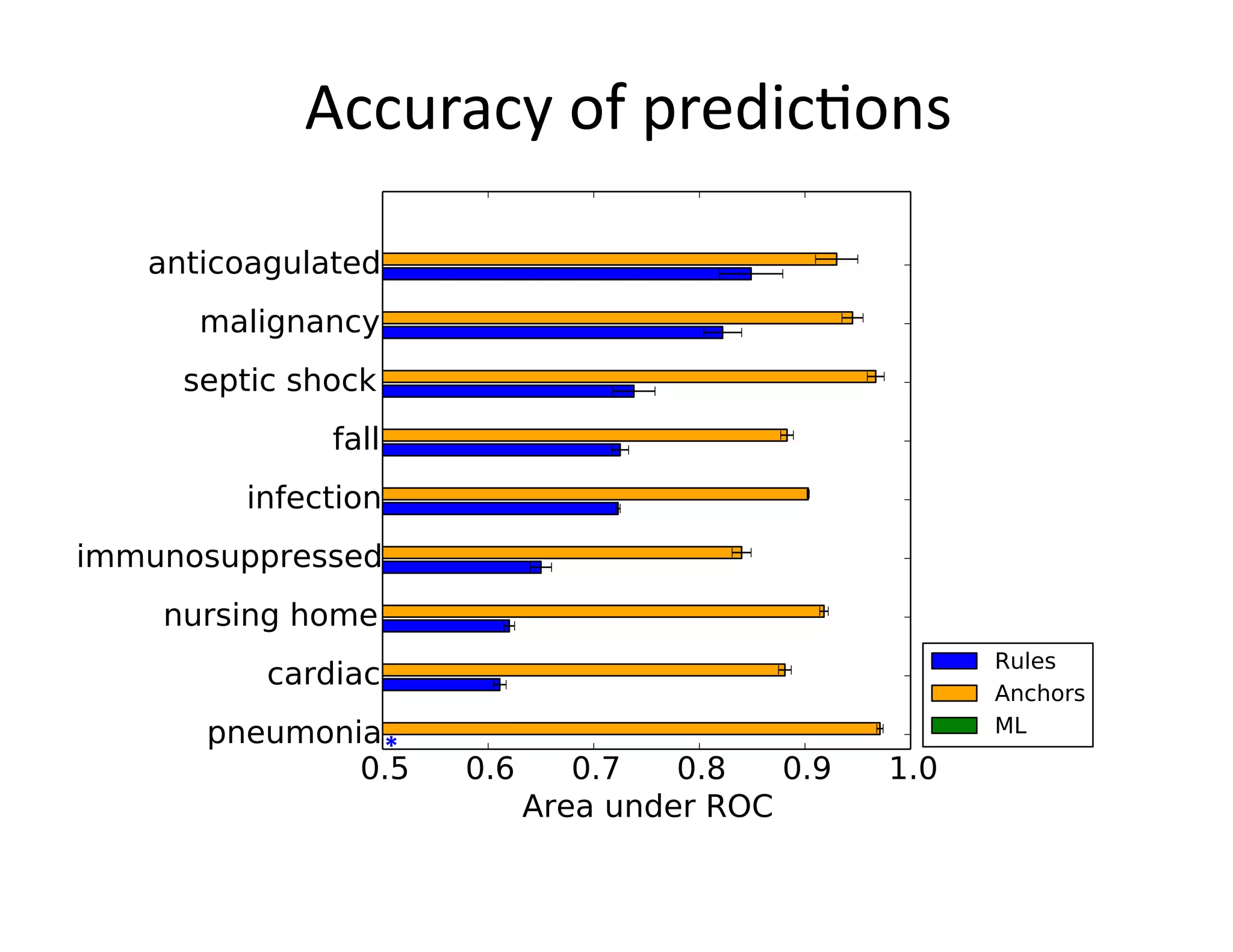
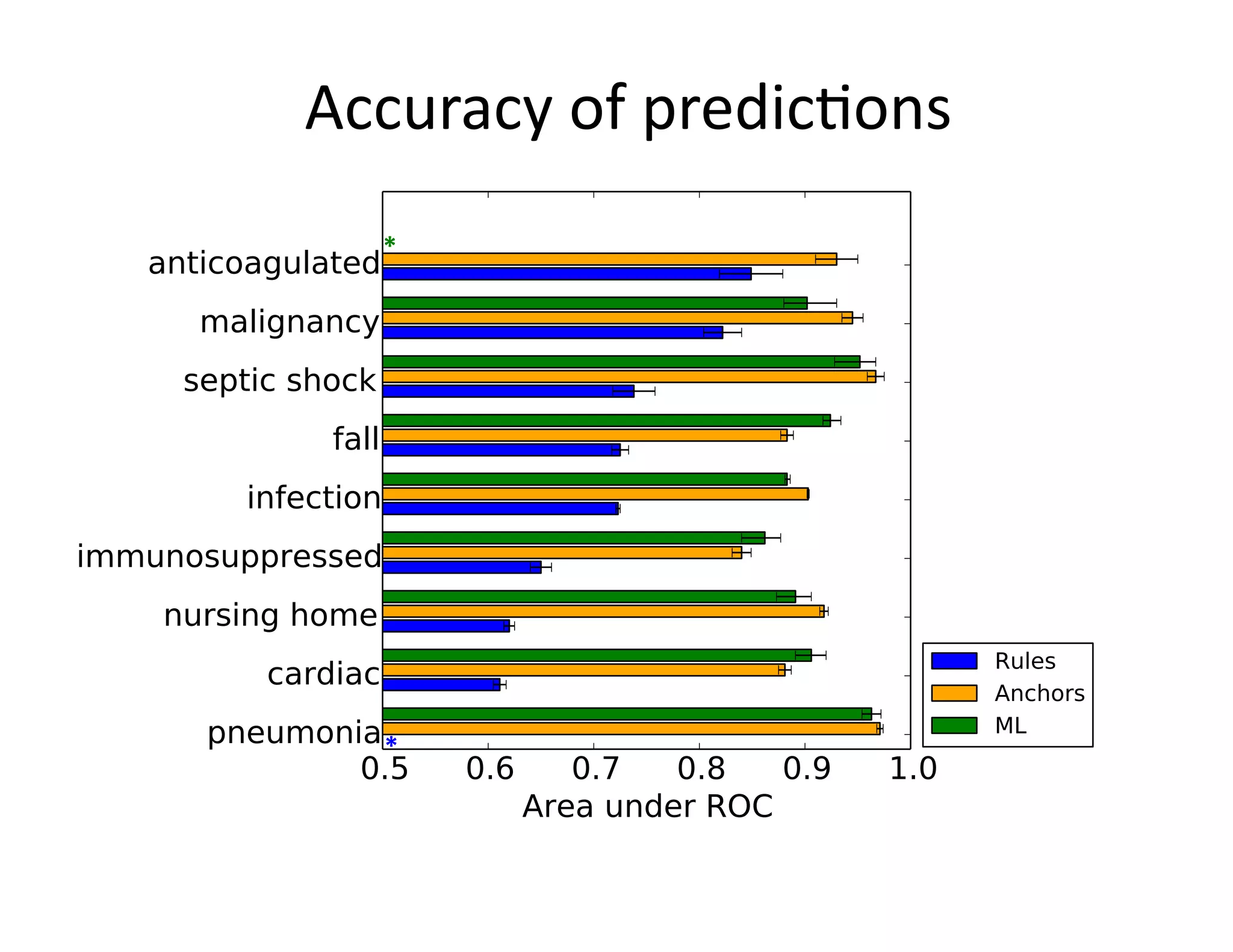
![Example:
Triggering
Clinical
Pathways
• Clinical
Pathways
project
at
Beth
Israel
Deaconess
Medical
Center
(BIDMC)
• Standardizing
care
in
the
Emergency
Department
– Reduce
possibili@es
for
error
– Enforce
established
best
prac@ces
• Pathways
have
been
shown
to
reduce
in-‐hospital
complica@ons,
without
increasing
costs
[Rober
et
al
2010]](https://image.slidesharecdn.com/ibmcognitivecomputingsept162015-150917131628-lva1-app6892/75/Using-Machine-Learning-to-Automate-Clinical-Pathways-12-2048.jpg)
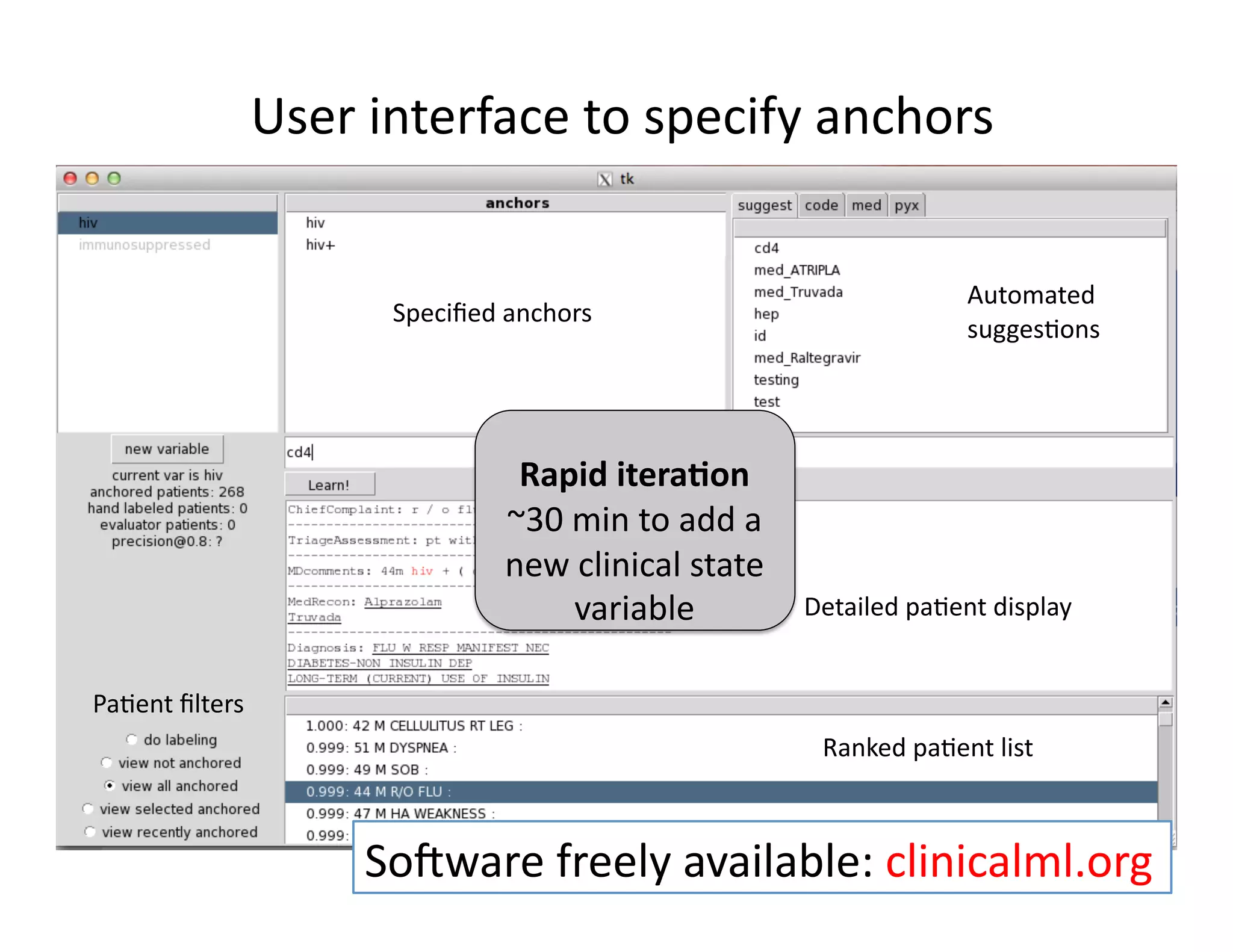
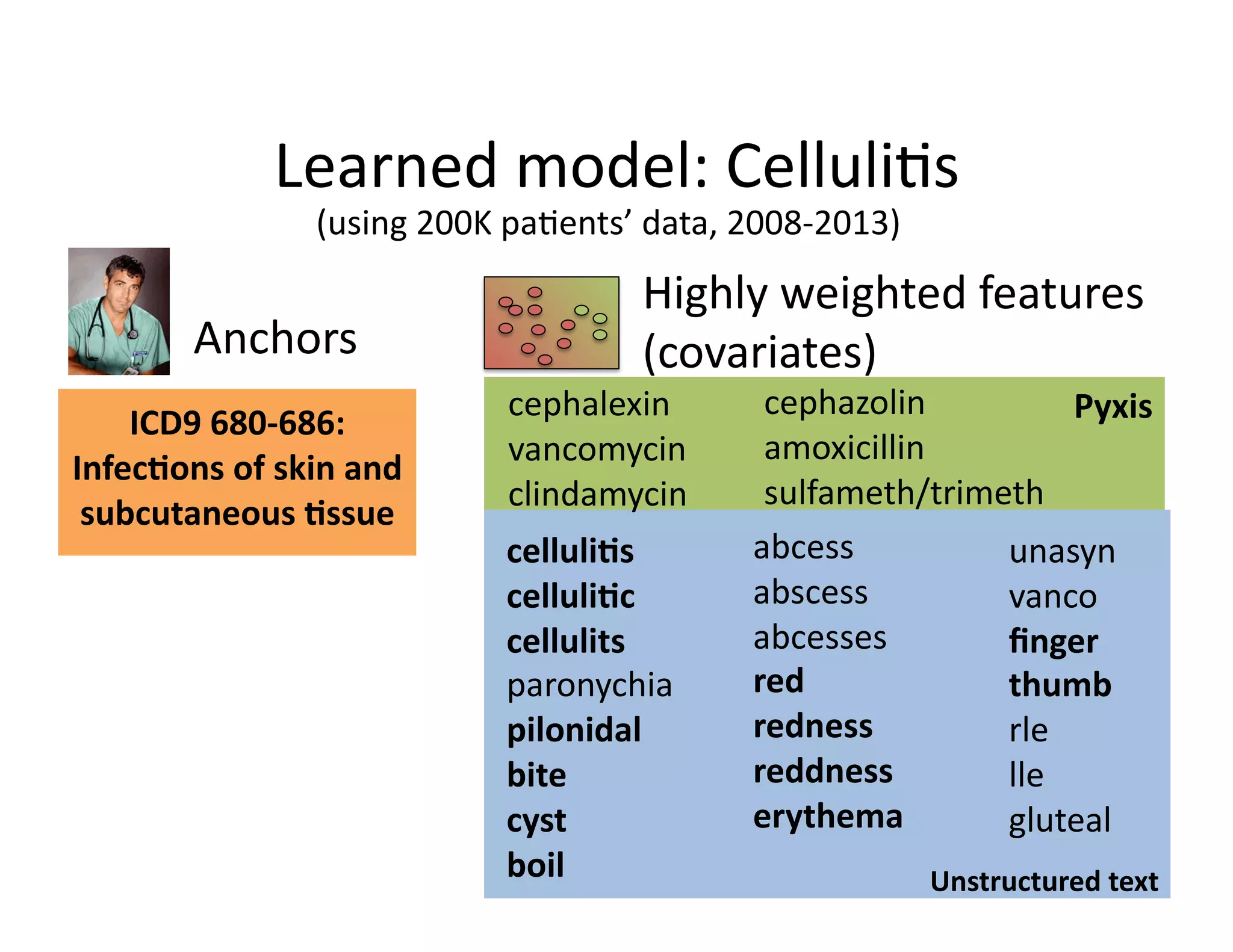
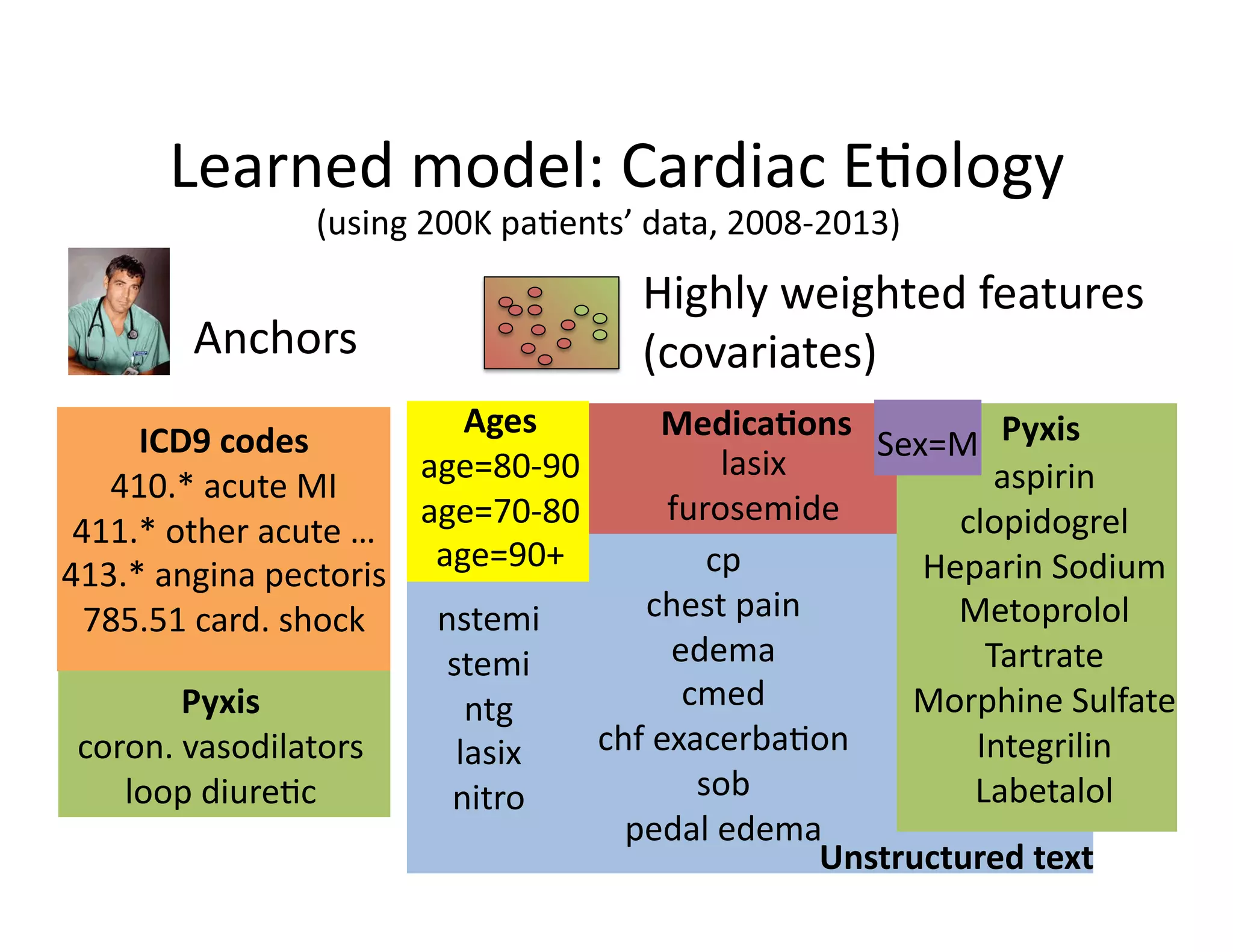
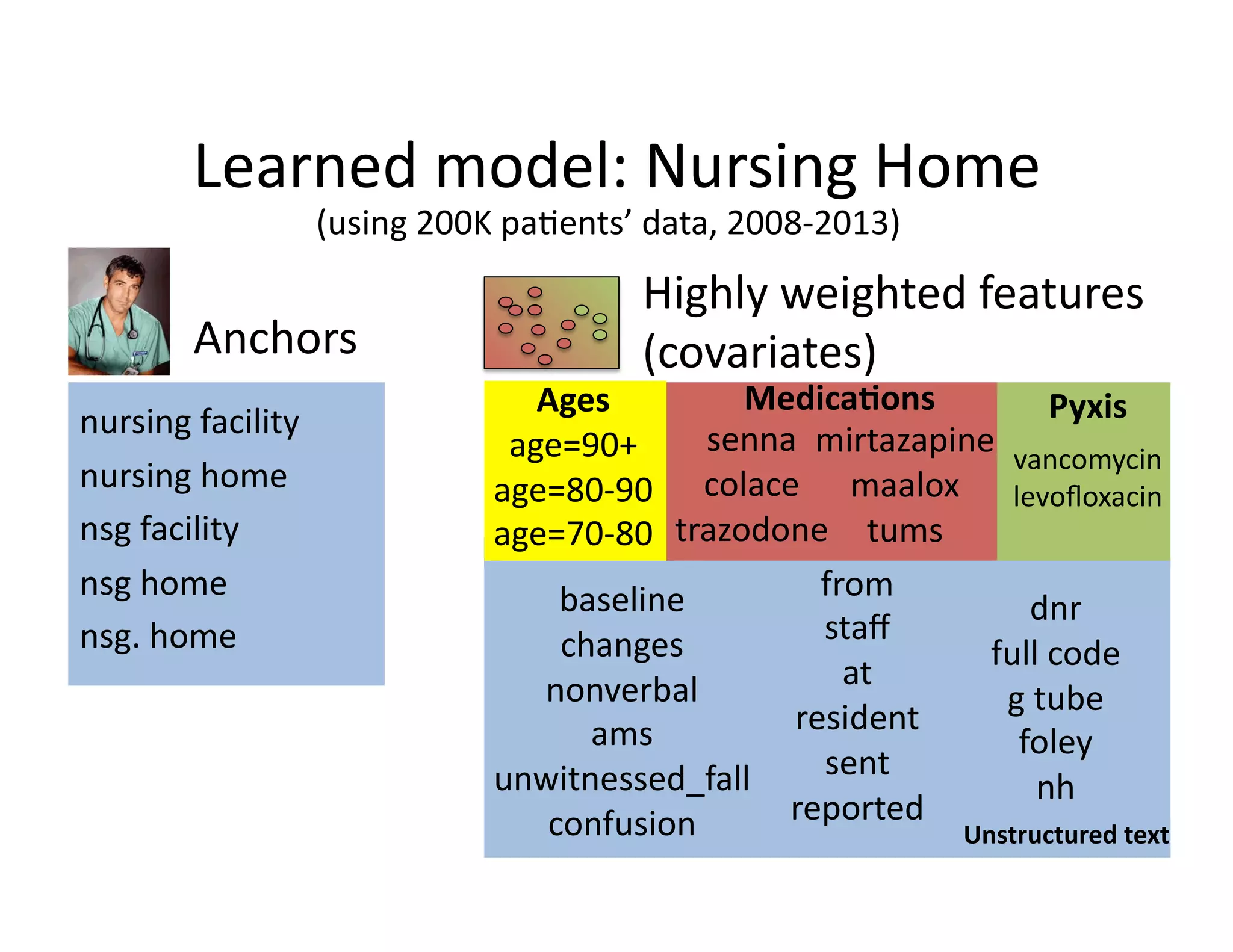
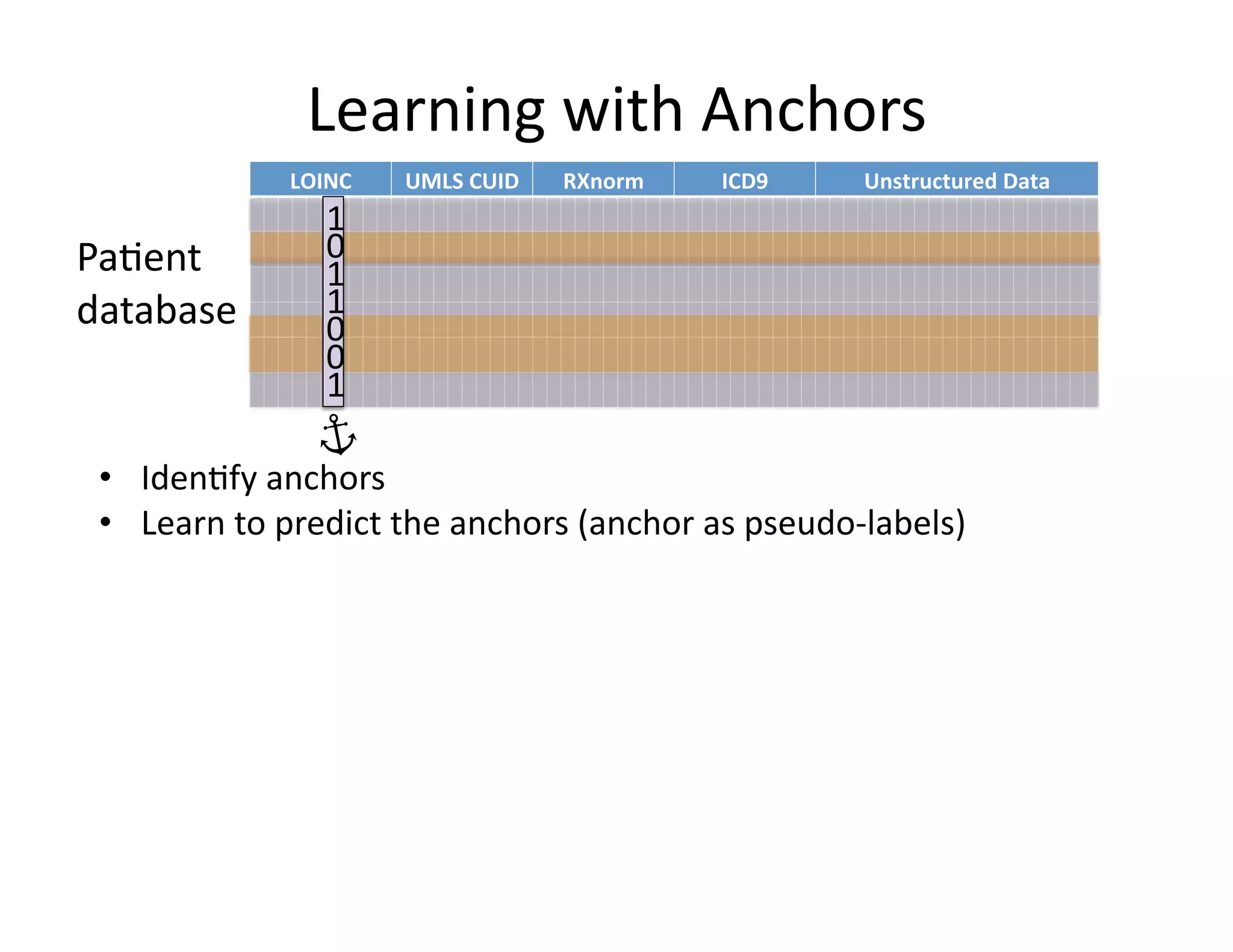
![Our
contribu@on:
Anchor
&
Learn
Framework
• Use
a
combina@on
of
domain
exper@se
(simple
rules)
and
vast
amounts
of
data
(machine
learning).
• Method
does
not
require
any
manual
labeling.
• Anchors
are
highly
transferable
between
ins@tu@ons.
[Halpern
et
al.,
AMIA
2014]](https://image.slidesharecdn.com/ibmcognitivecomputingsept162015-150917131628-lva1-app6892/75/Using-Machine-Learning-to-Automate-Clinical-Pathways-18-2048.jpg)

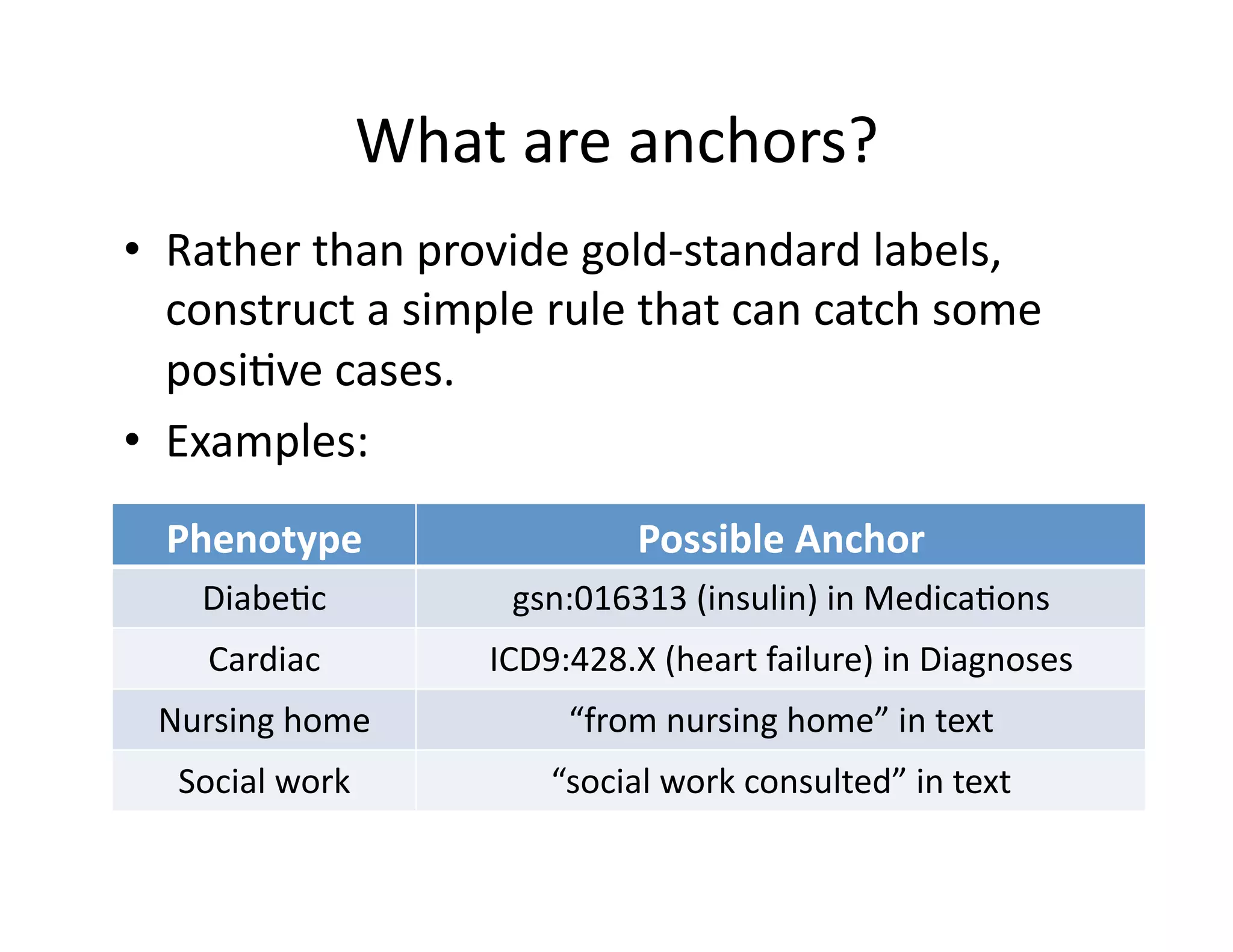
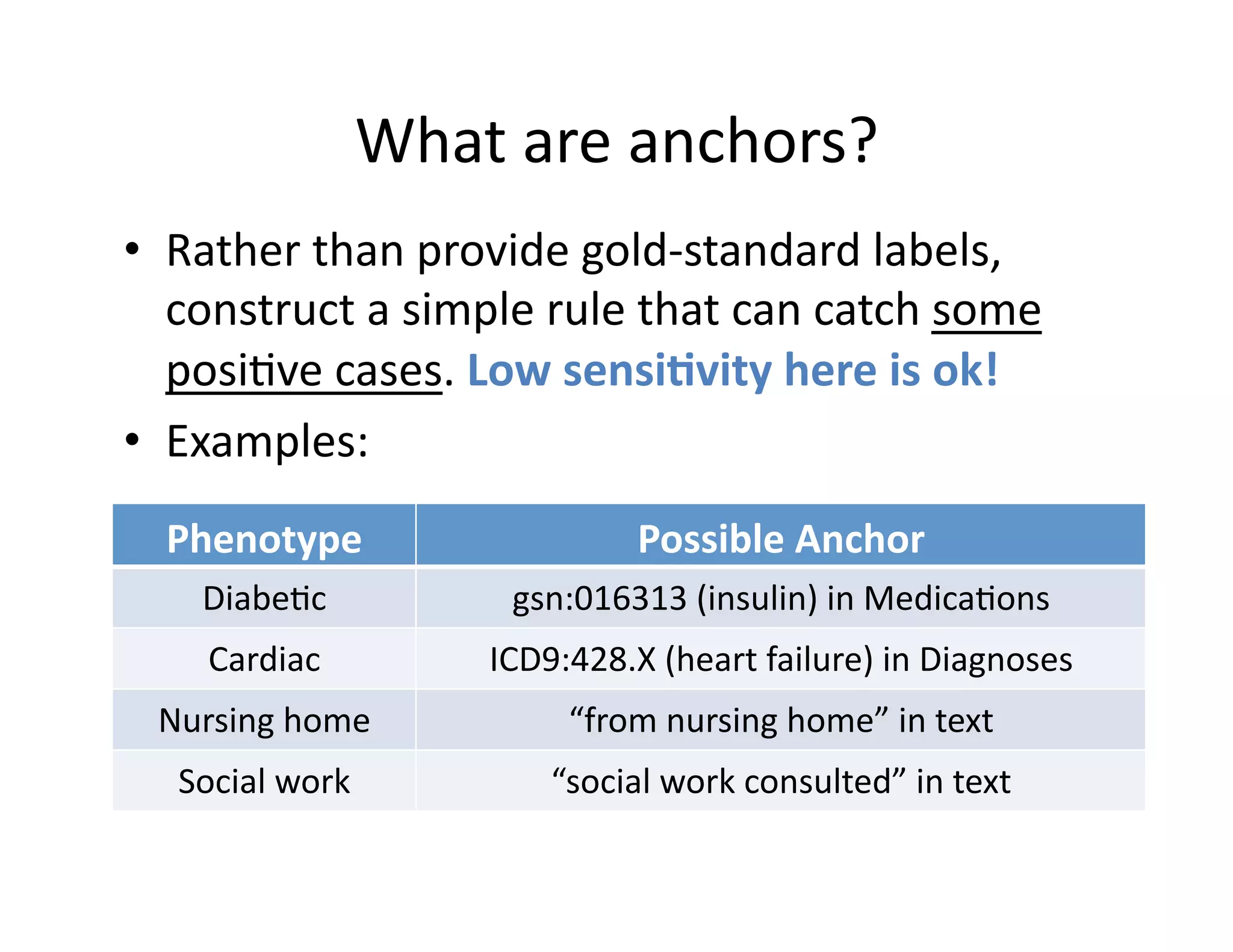

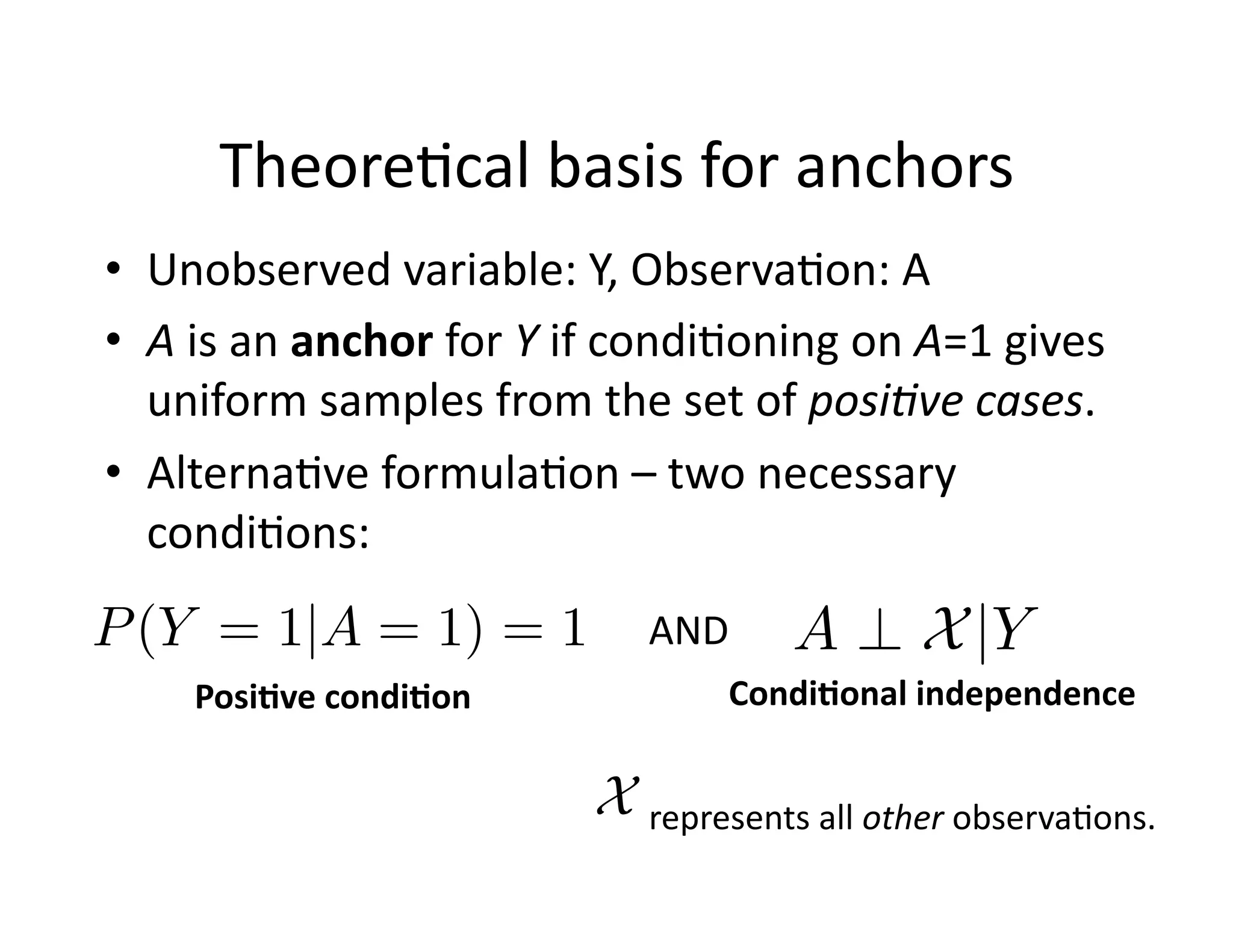
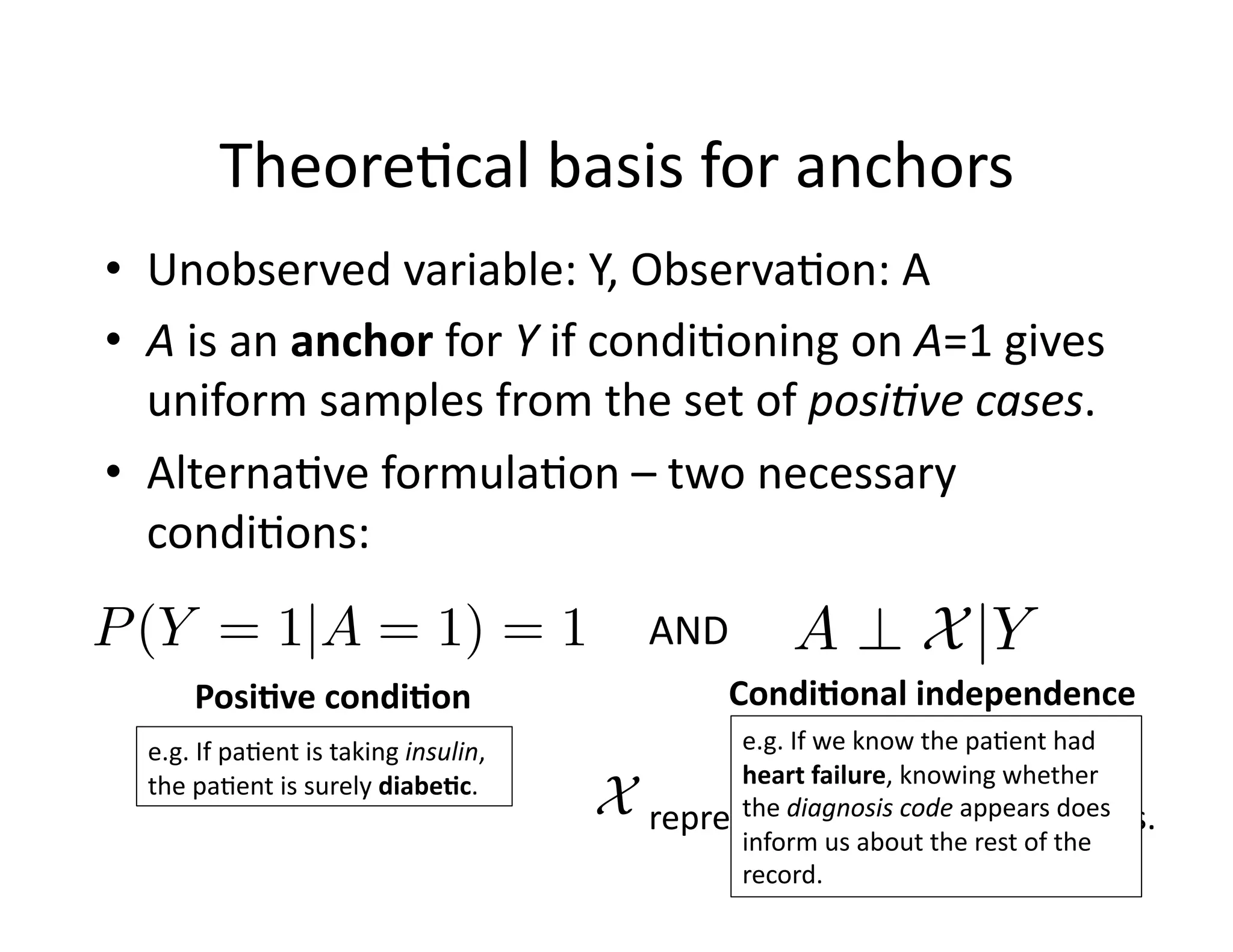
![Theore@cal
basis
for
anchors
• Unobserved
variable:
Y,
Observa@on:
A
• A
is
an
anchor
for
Y
if
condi@oning
on
A=1
gives
uniform
samples
from
the
set
of
posi8ve
cases.
• Theorem
[Elkan
&
Noto
2008]:
In
the
above
se>ng,
a
func8on
to
predict
A
can
be
transformed
to
predict
Y
• Can
also
use
more
recent
advances
on
learning
with
noisy
labels
(e.g.,
Natarajan
et
al.,
NIPS
‘13)](https://image.slidesharecdn.com/ibmcognitivecomputingsept162015-150917131628-lva1-app6892/75/Using-Machine-Learning-to-Automate-Clinical-Pathways-34-2048.jpg)
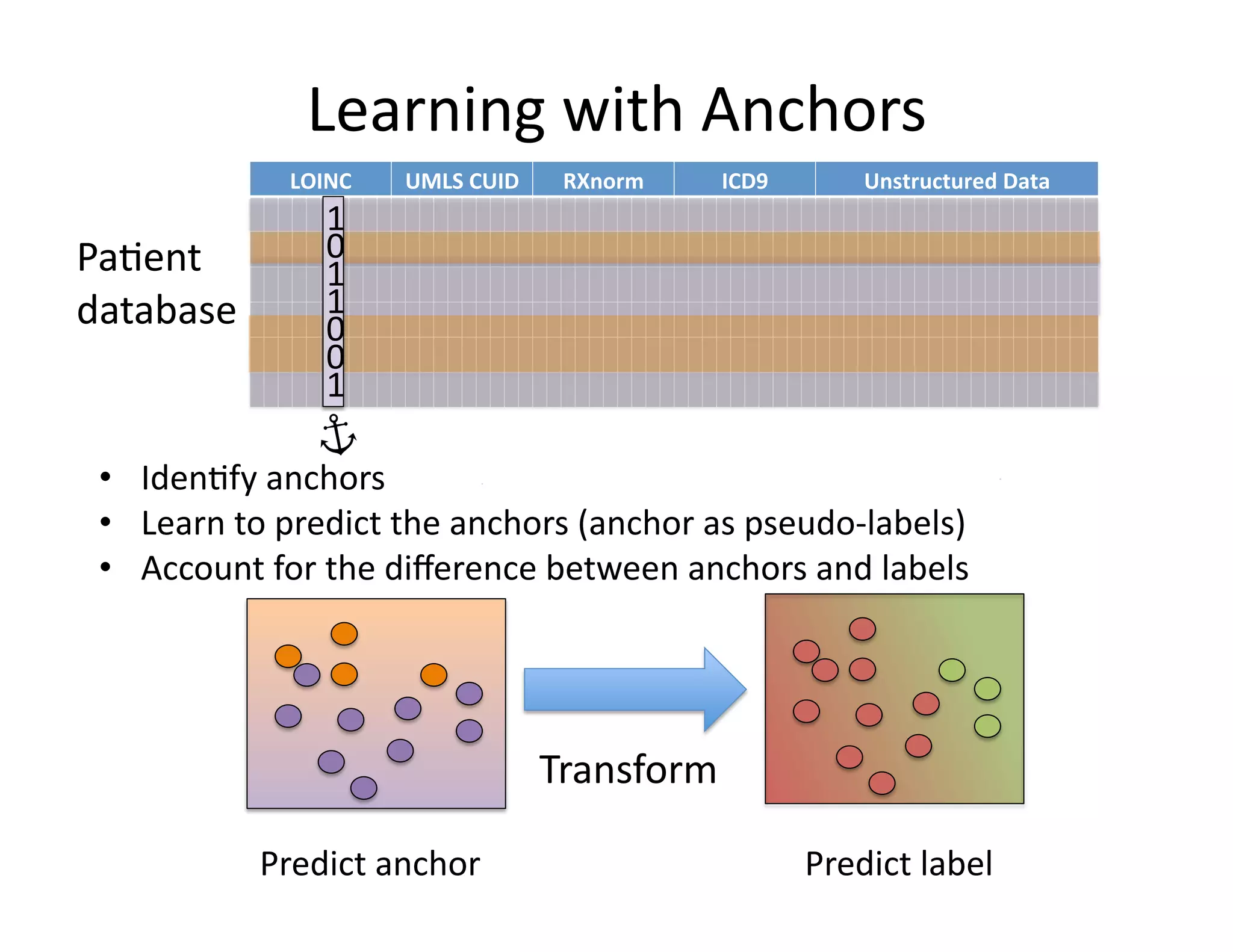
![Learning
with
anchors
Input:
anchor
A
unlabeled
pa@ents
Output:
predic@on
rule
1. Learn
a
calibrated
classifier
(e.g.
logis@c
regression)
to
predict:
2. Using
a
validate
set,
let
P
be
the
pa@ents
with
A=1.
Compute:
3. For
a
previously
unseen
pa@ent
t,
predict:
Pr(A = 1 | ˜X)
C =
1
|P|
X
k2P
Pr(A = 1 | ˜X(k)
)
[Elkan
&
Noto
2008]
1
C
Pr(A = 1|X(t)
) if A(t)
= 0
1 if A(t)
= 1
Calibra*on
C
is
the
average
model
predic@on
for
pa@ents
with
anchors.
Learning
Learn
to
predict
A
from
the
other
variables.
Transforma*on
If
no
anchor
present,
according
to
a
scaled
version
of
the
anchor-‐predic@on
model.](https://image.slidesharecdn.com/ibmcognitivecomputingsept162015-150917131628-lva1-app6892/75/Using-Machine-Learning-to-Automate-Clinical-Pathways-35-2048.jpg)