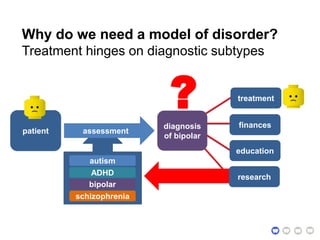
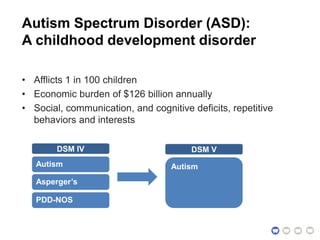
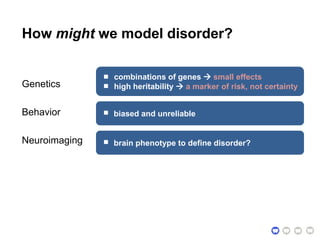
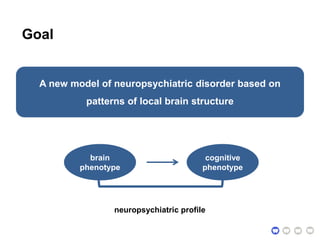
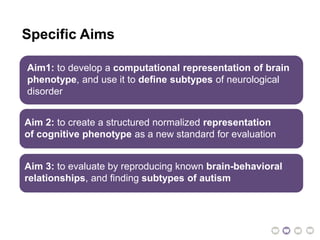
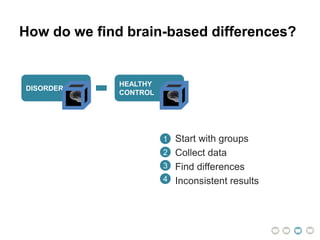
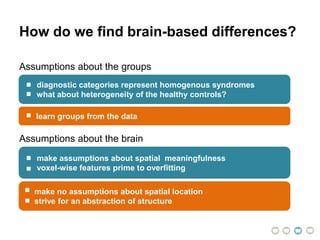
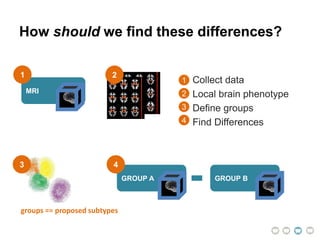
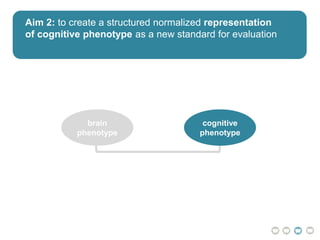
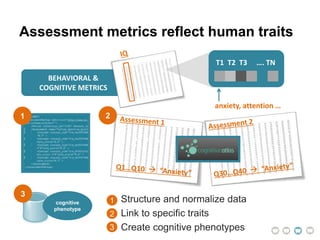
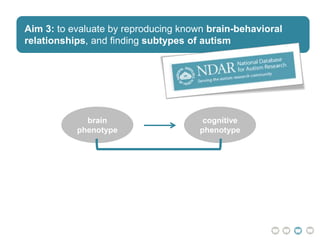
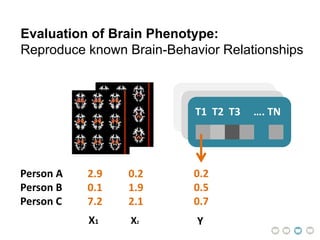
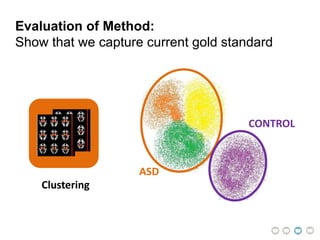
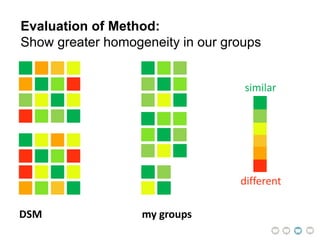
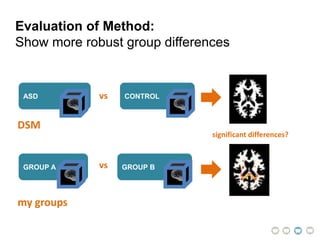
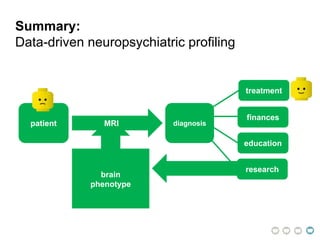
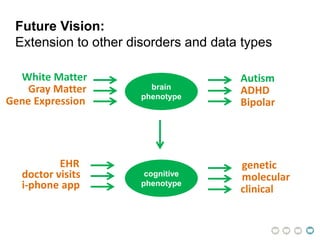
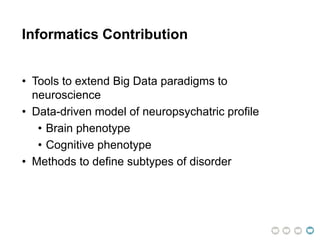
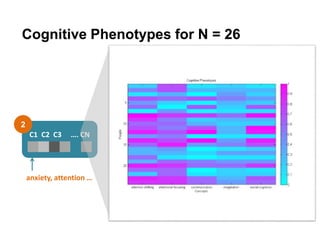
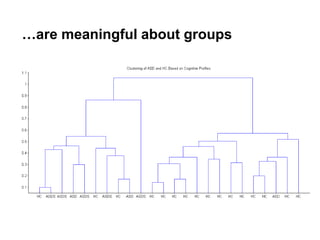
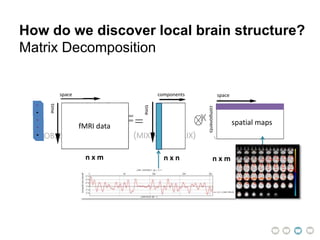
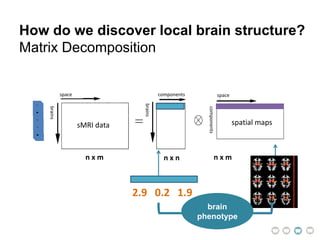
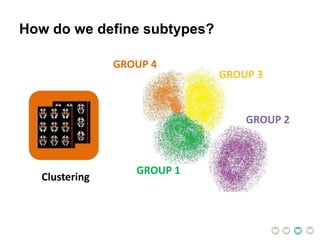
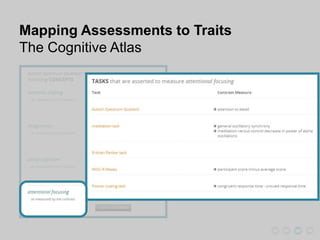
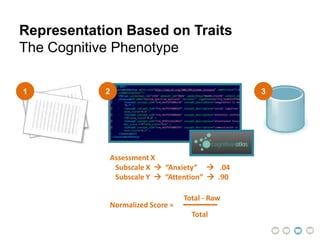
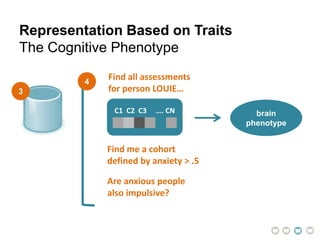
This document presents a proposal for developing a data-driven model of neuropsychiatric disorders based on analyzing patterns in brain structure and cognitive assessments. The goals are to define clinically useful subtypes of disorders, create standardized representations of brain and cognitive phenotypes, and evaluate the model by reproducing known relationships between the brain and behavior. Matrix decomposition would be used to discover local brain structure from neuroimaging data. Clustering would define disorder subtypes. Cognitive assessments would be mapped to traits and normalized to create cognitive phenotypes. The model could help enable personalized treatment by discovering biomarkers of disorders.