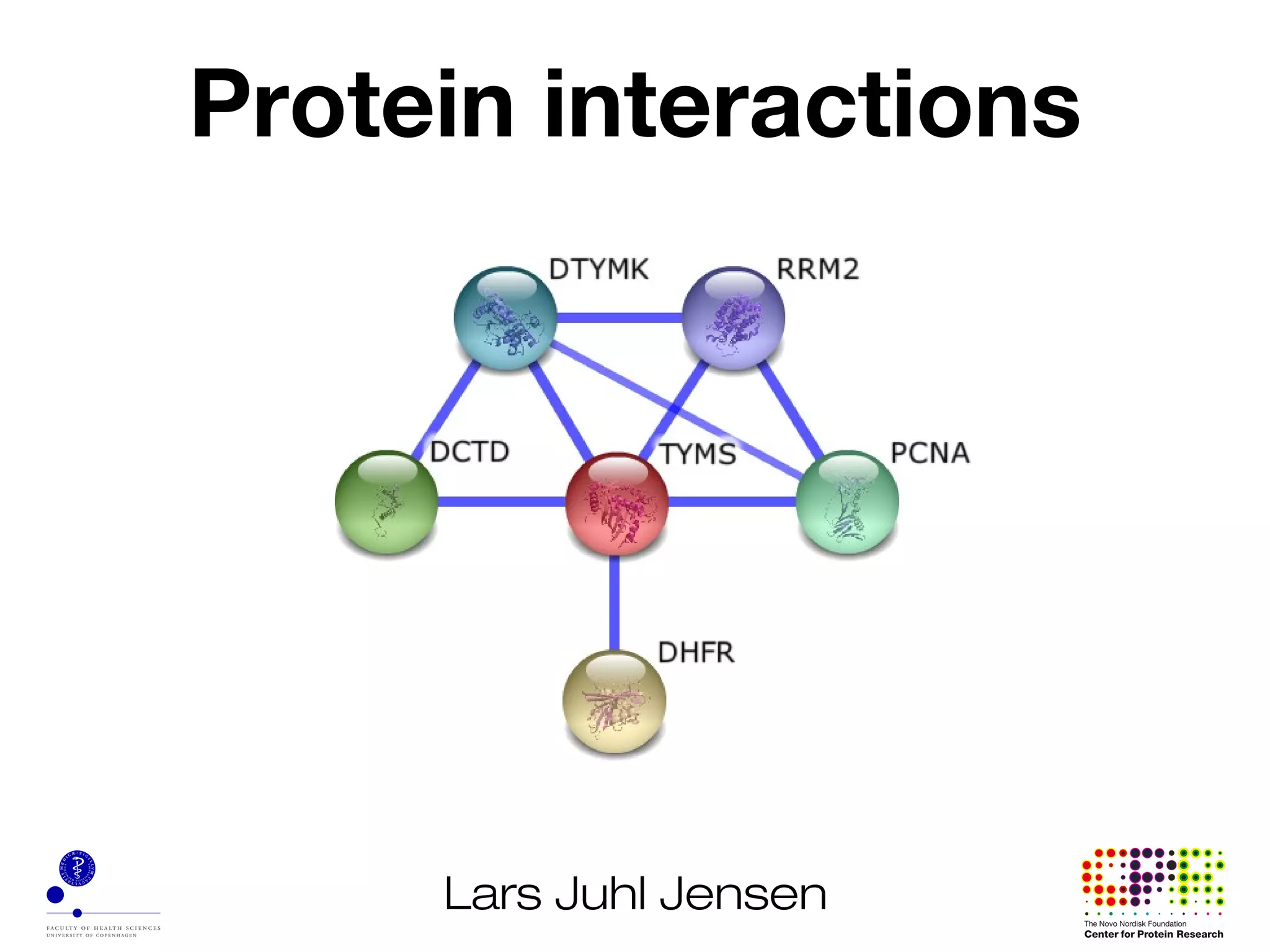
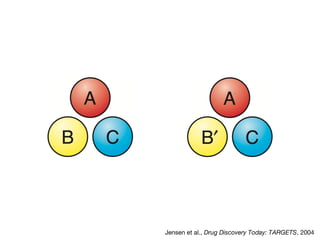
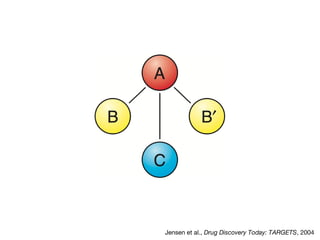
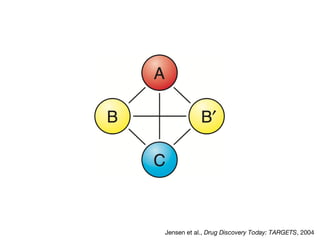
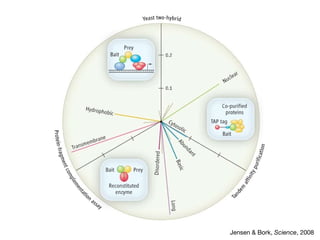
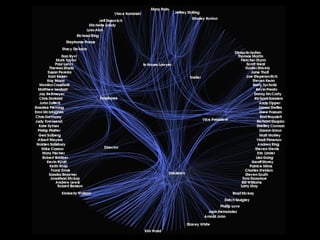
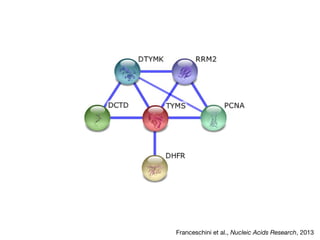
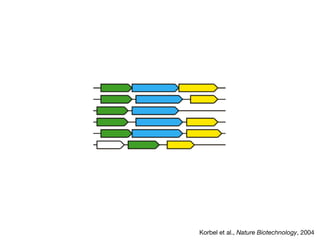
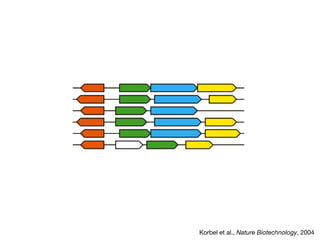
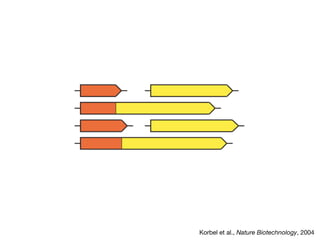
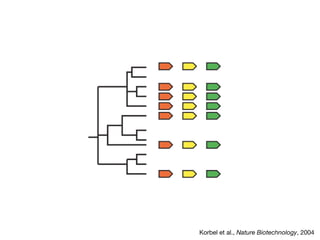
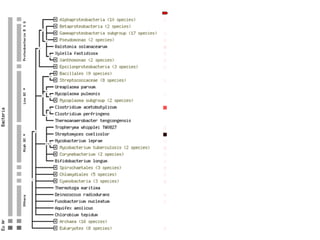
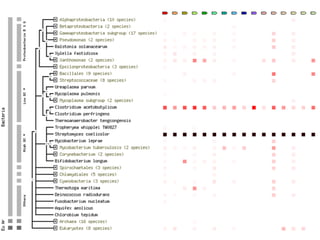
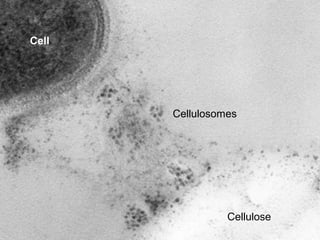
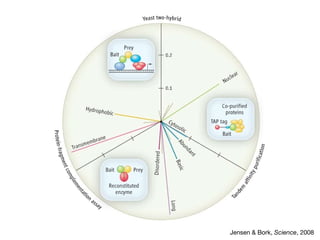
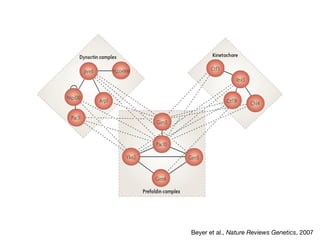
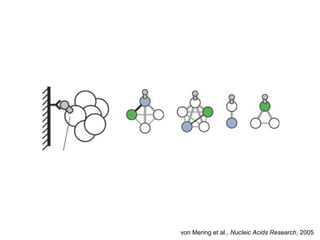
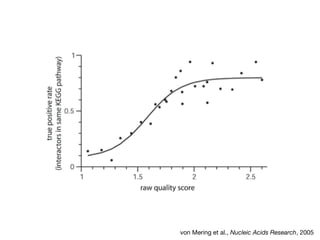
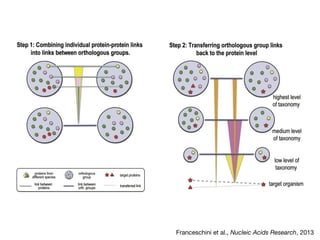
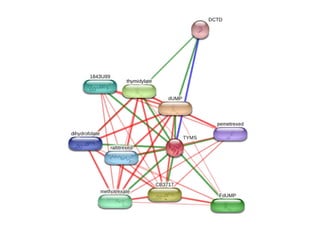
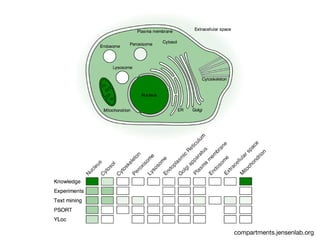
This document discusses protein-protein interaction data and resources. It introduces common interaction detection methods like yeast two-hybrid and affinity purification. Several curated interaction databases are described, including BioGRID, DIP, IntAct, and MINT. The document also discusses the STRING database, which integrates interaction data with genomic context predictions. It notes challenges like errors, biased assays, different formats between databases, and difficulty in comparing data across species. Quality scores and calibration to gold standards are presented as ways to assess confidence in interactions. The related STITCH database is also introduced, which integrates protein-protein and chemical-protein interactions.