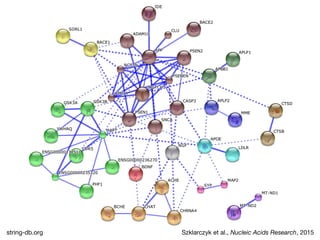
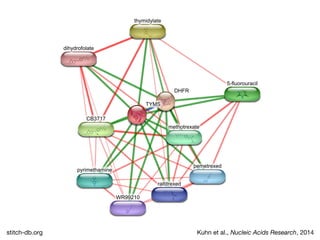
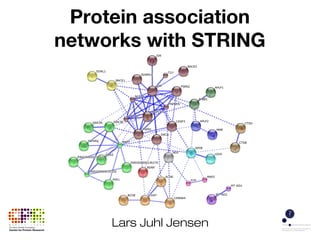
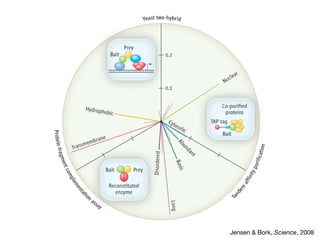
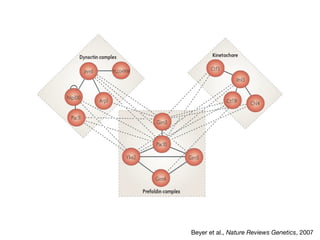
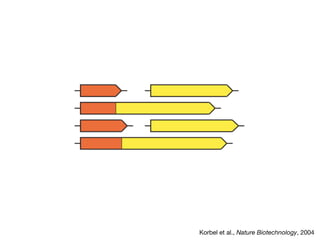
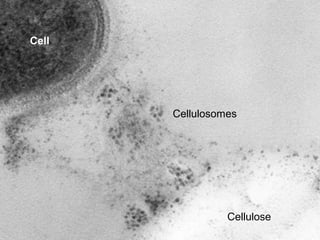
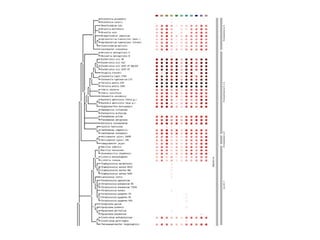
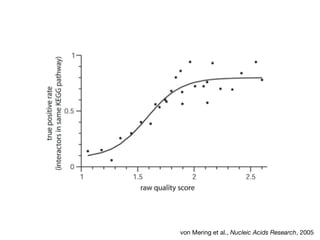
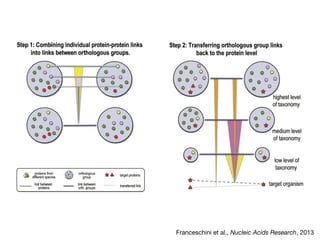
The document discusses the STRING protein association network database. It provides information from multiple sources, including curated databases, experiments, and predictions, to assign confidence scores and generate protein interaction networks. The document guides the reader through exercises exploring the human insulin receptor network on STRING, examining the evidence supporting its interaction with IRS1 and how confidence view and cutoffs alter the network displayed.











































































![Gene and protein names
Cue words for entity
recognition
Verbs for relation extraction
[nxexpr The expression of
[nxgene the cytochrome
genes
[nxpg CYC1 and CYC7]]]
is controlled by
[nxpg HAP1]
Saric et al., Proceedings of ACL, 2004](https://image.slidesharecdn.com/jensen2016talk8-160619152956/85/Protein-association-networks-with-STRING-101-320.jpg)