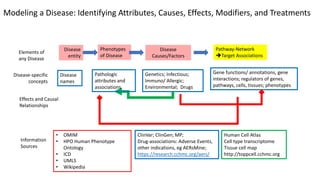
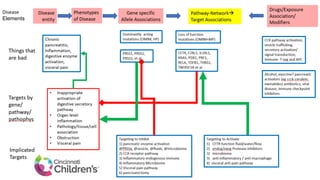
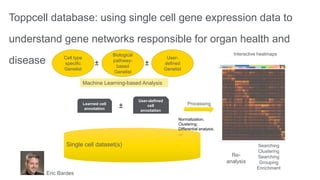
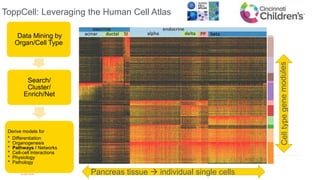
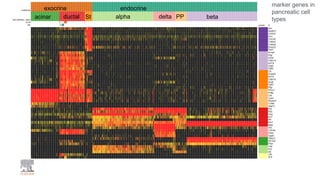
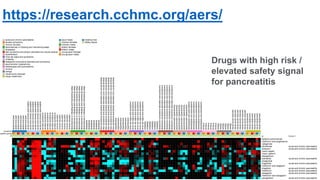
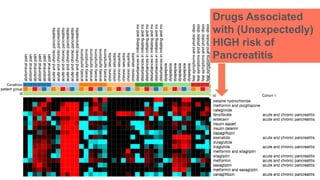
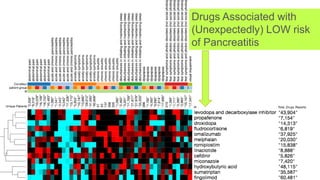
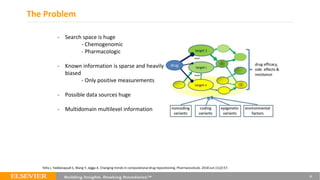
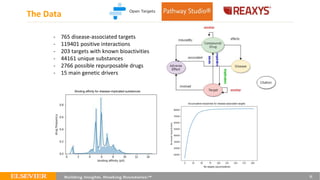
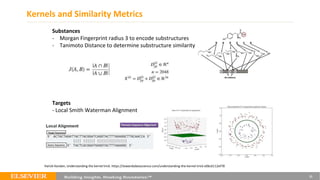
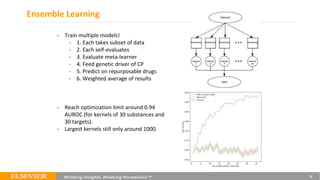
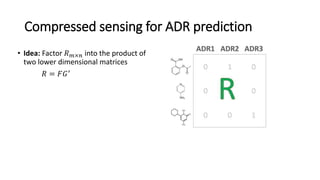
This document summarizes a webinar on using machine learning and data mining techniques to predict drug repurposing opportunities for chronic pancreatitis. Specifically:
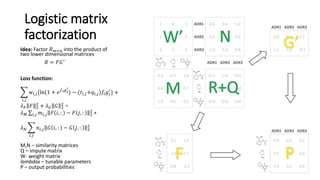
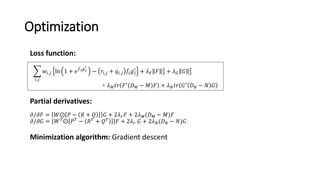
1. Ensemble learning techniques like kernel-based models were used to analyze drug and disease target interaction data from multiple sources to identify potential drug candidates for repurposing.
2. The top 5 repurposing candidates identified through this process were being evaluated further by the partner organization Mission-Cure with the goal of beginning patient trials by January 2020.
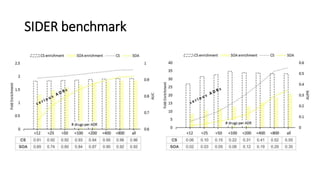
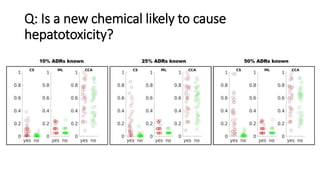
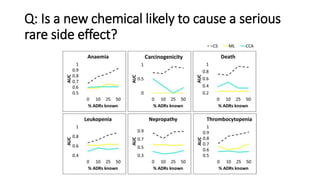
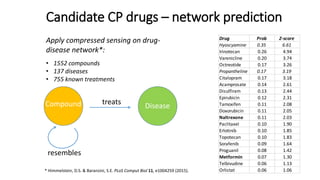
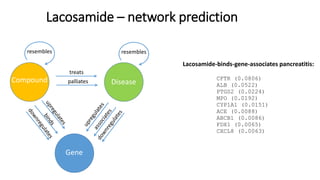
3. Additional techniques discussed included using compressed sensing to analyze drug-disease networks and predict side effects to help evaluate candidate drugs identified for repurposing opportunities.