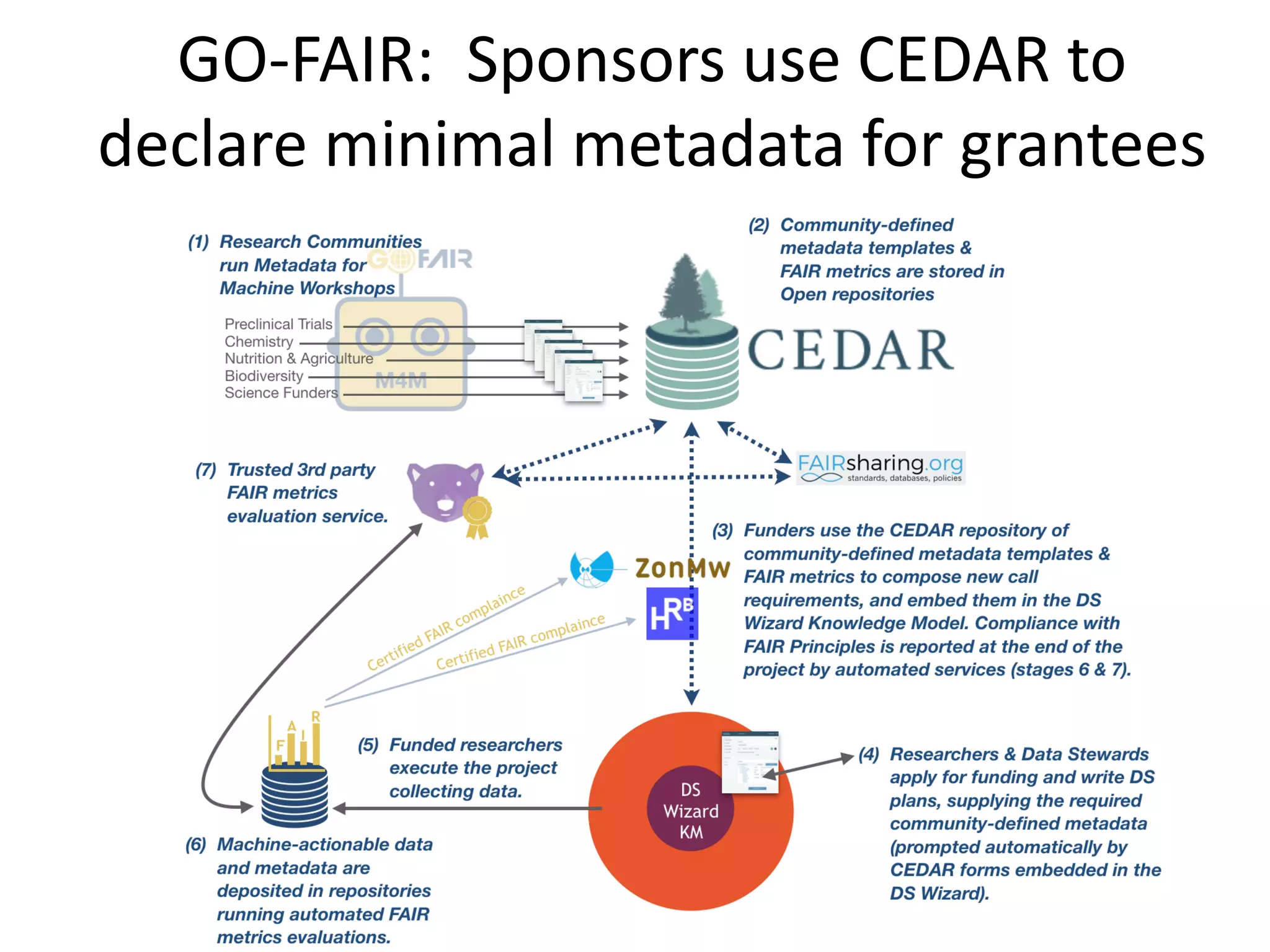
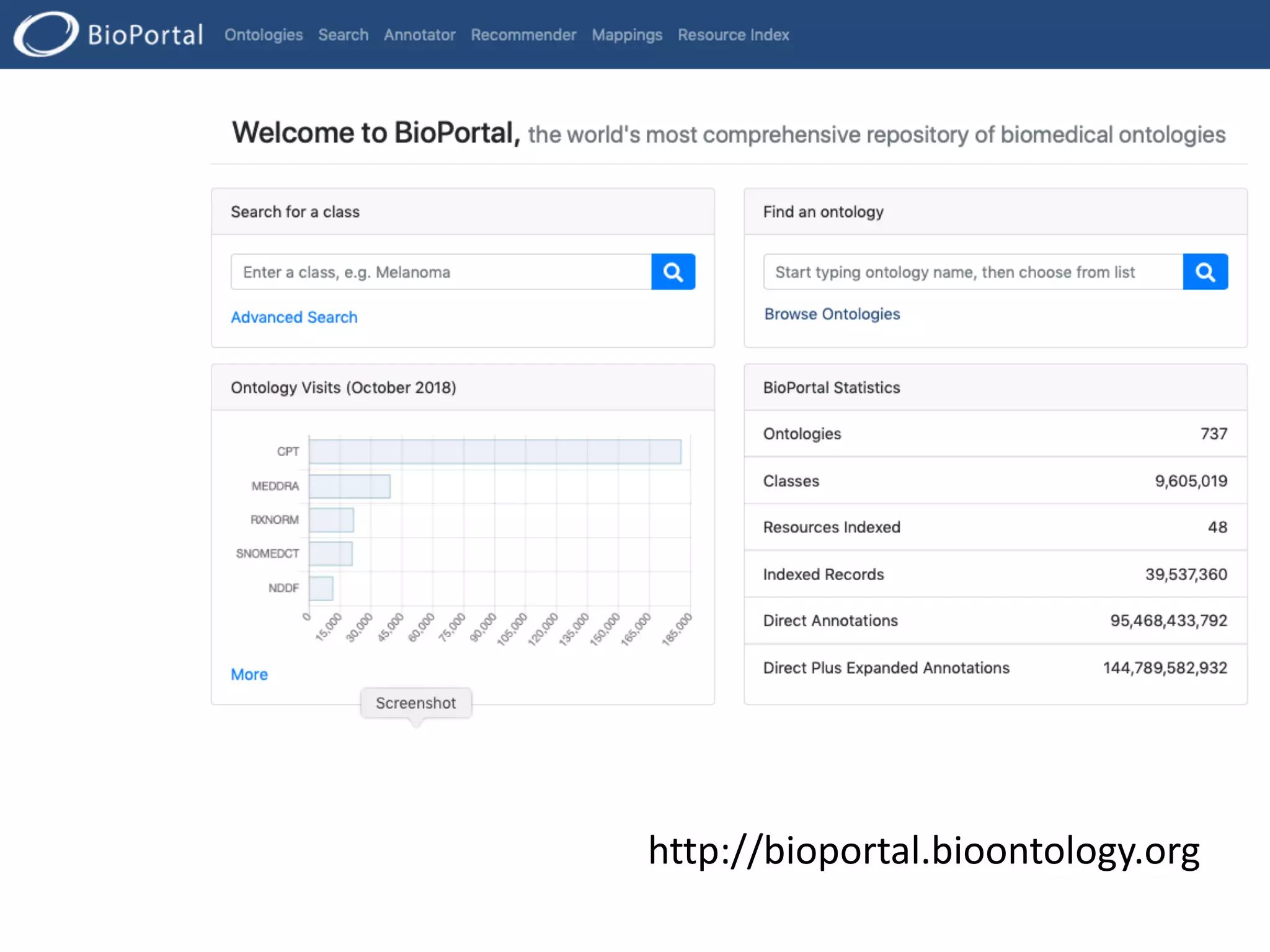
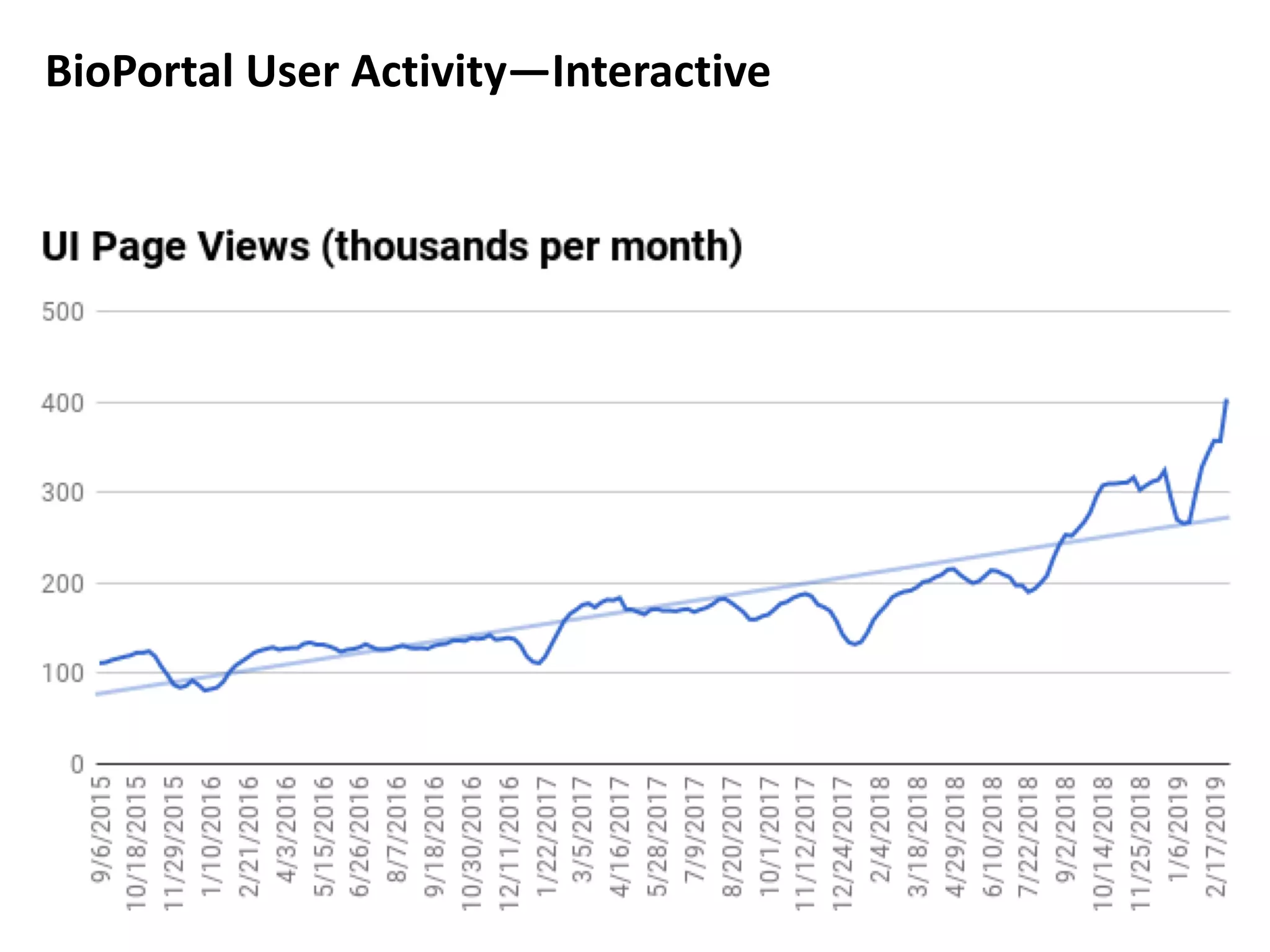
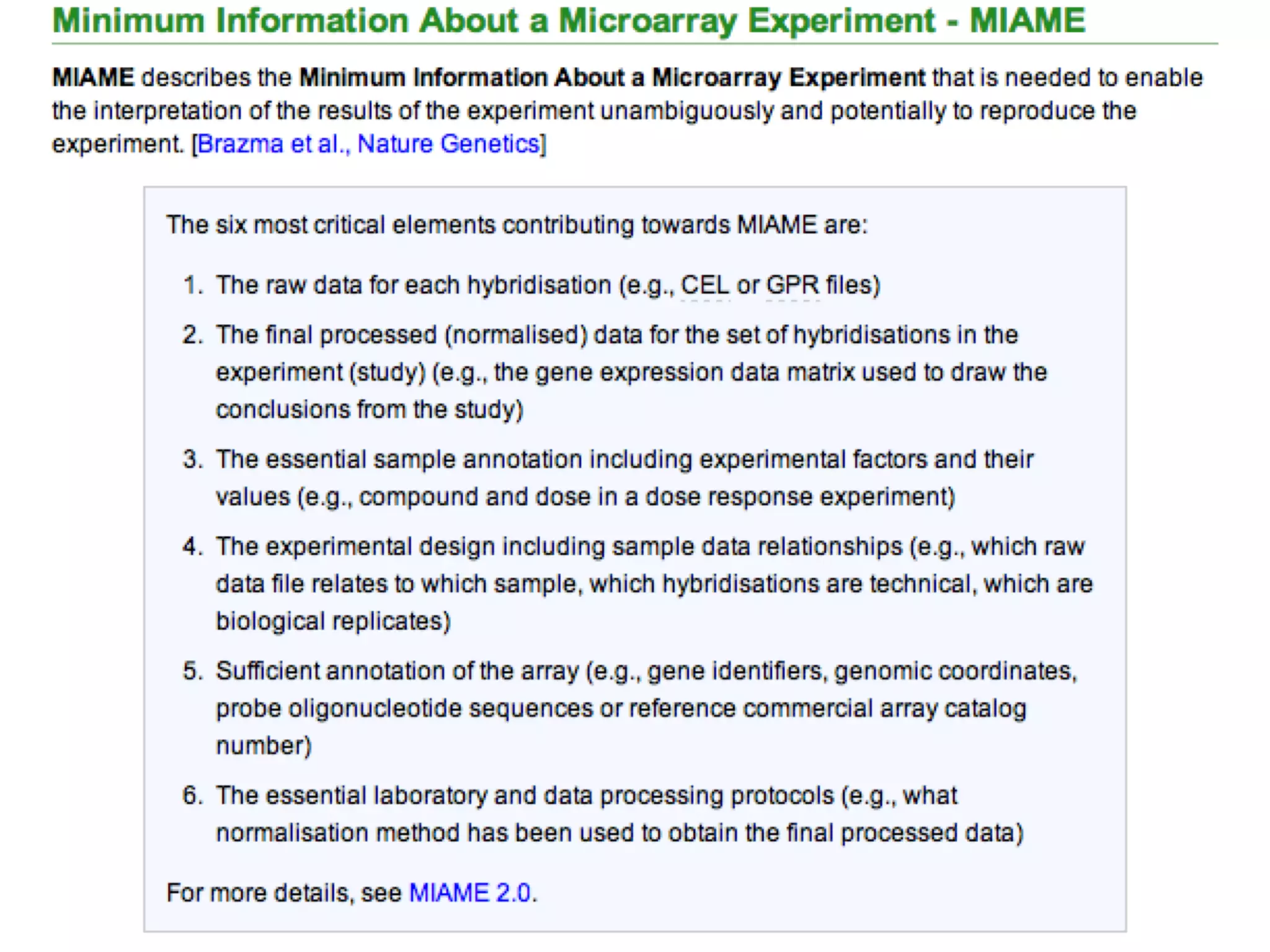
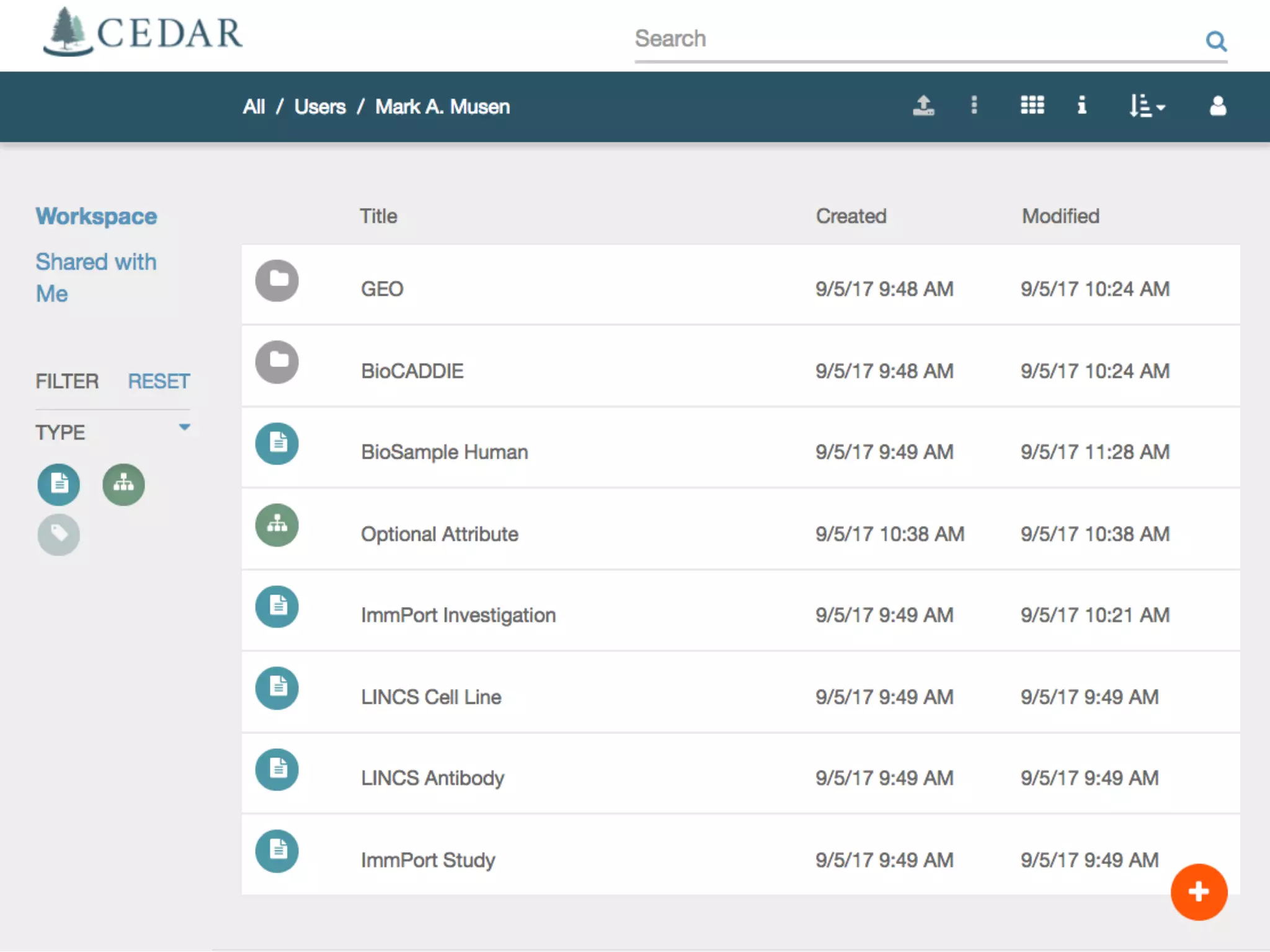
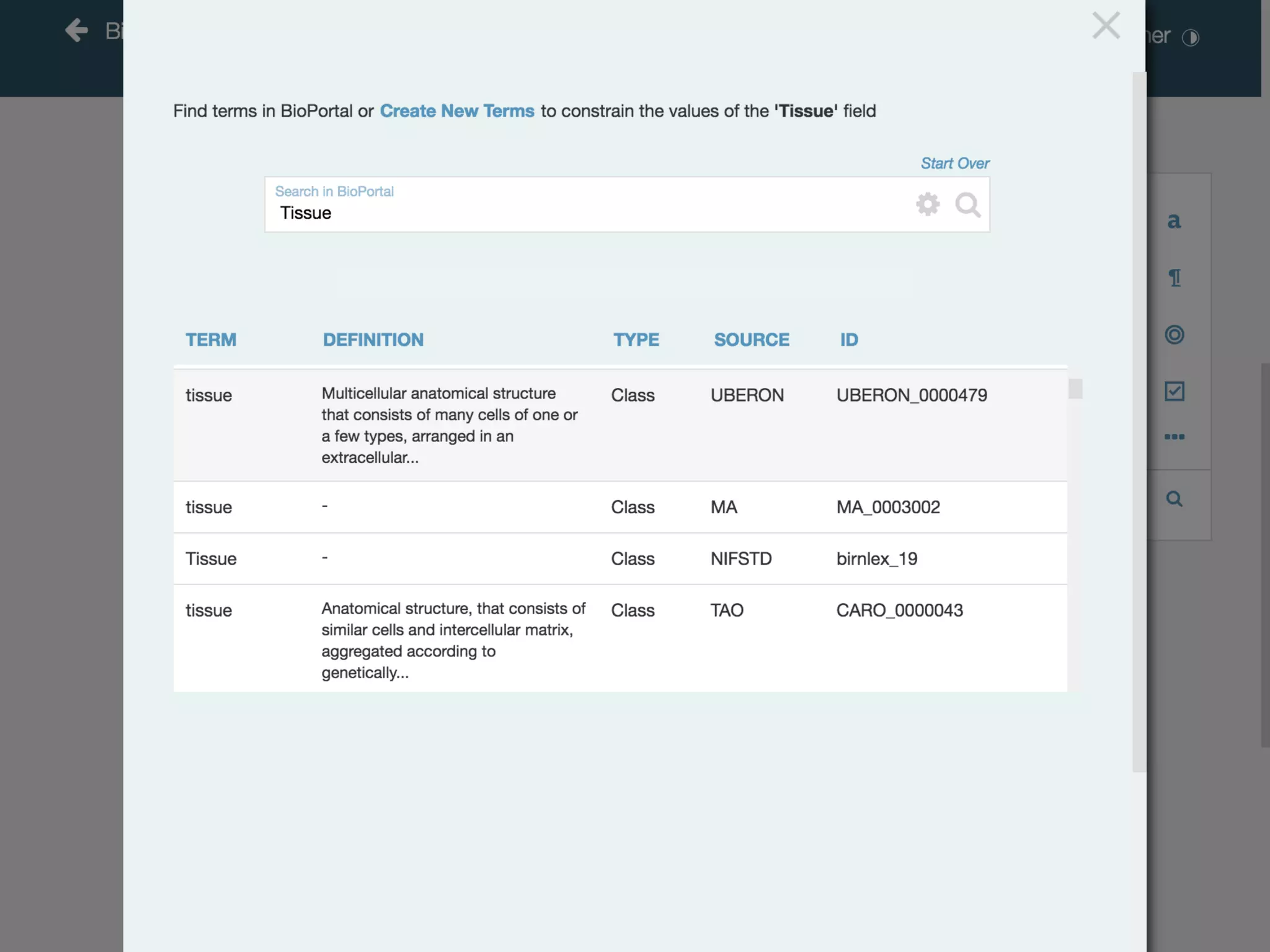
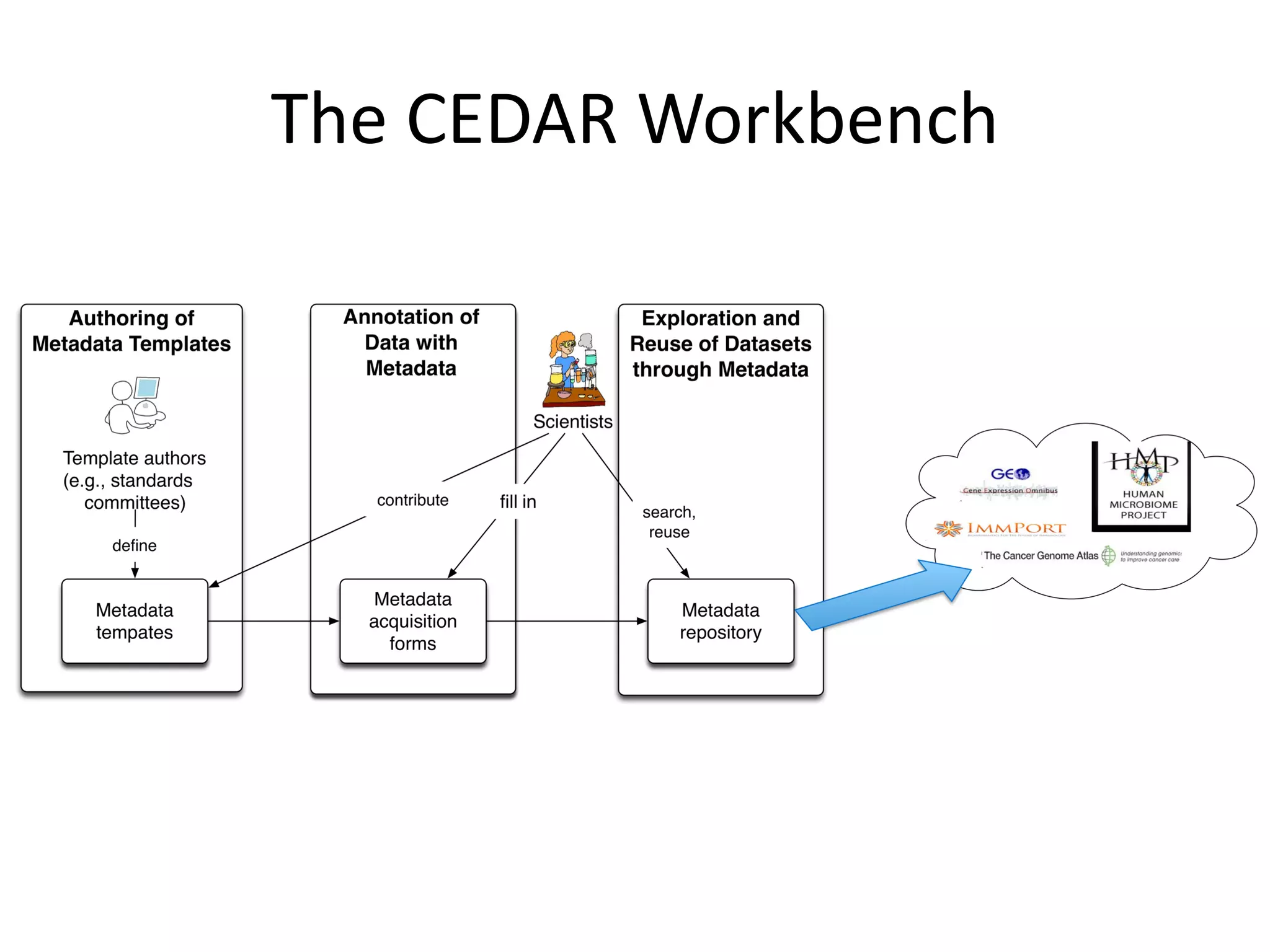
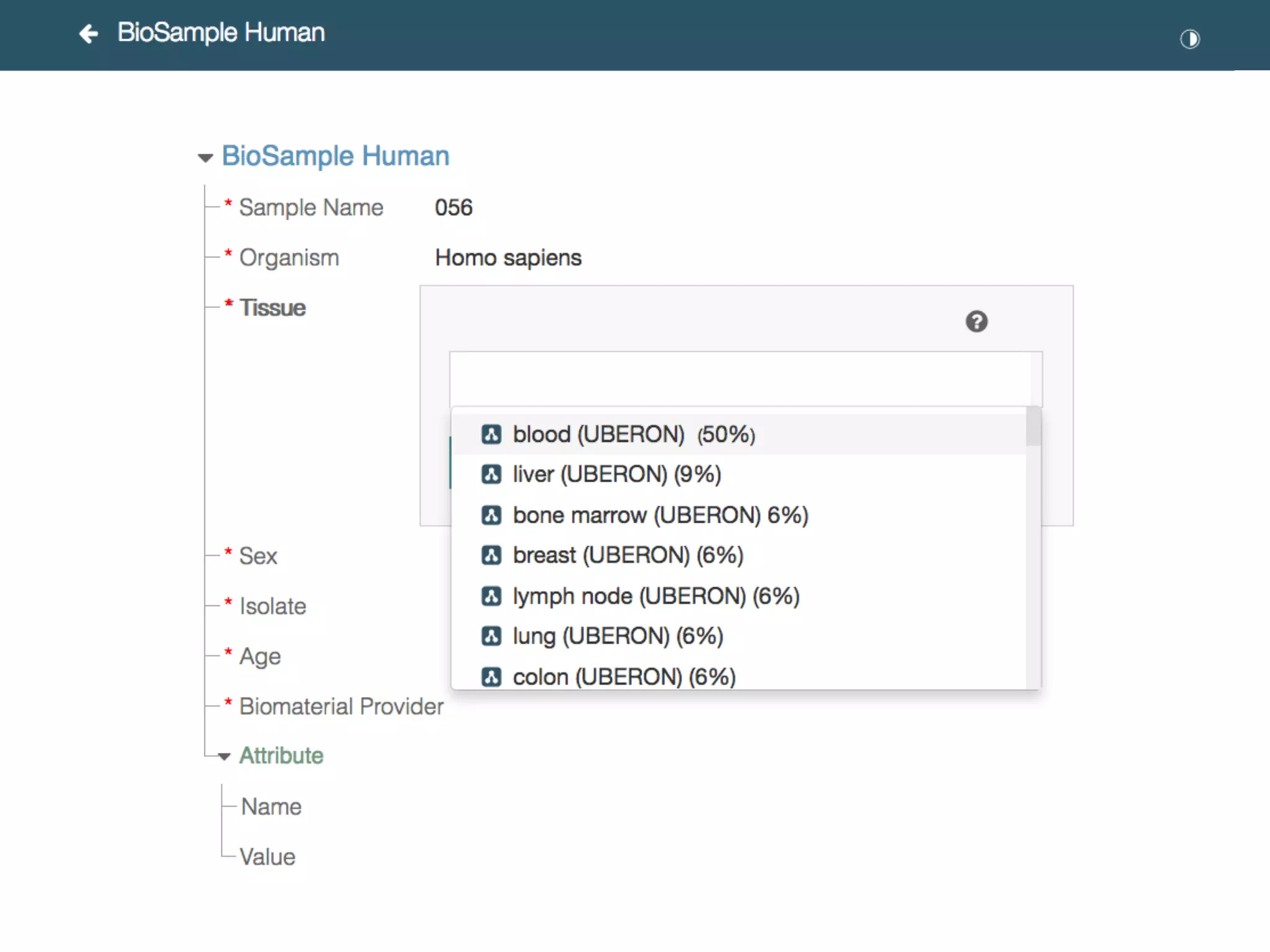
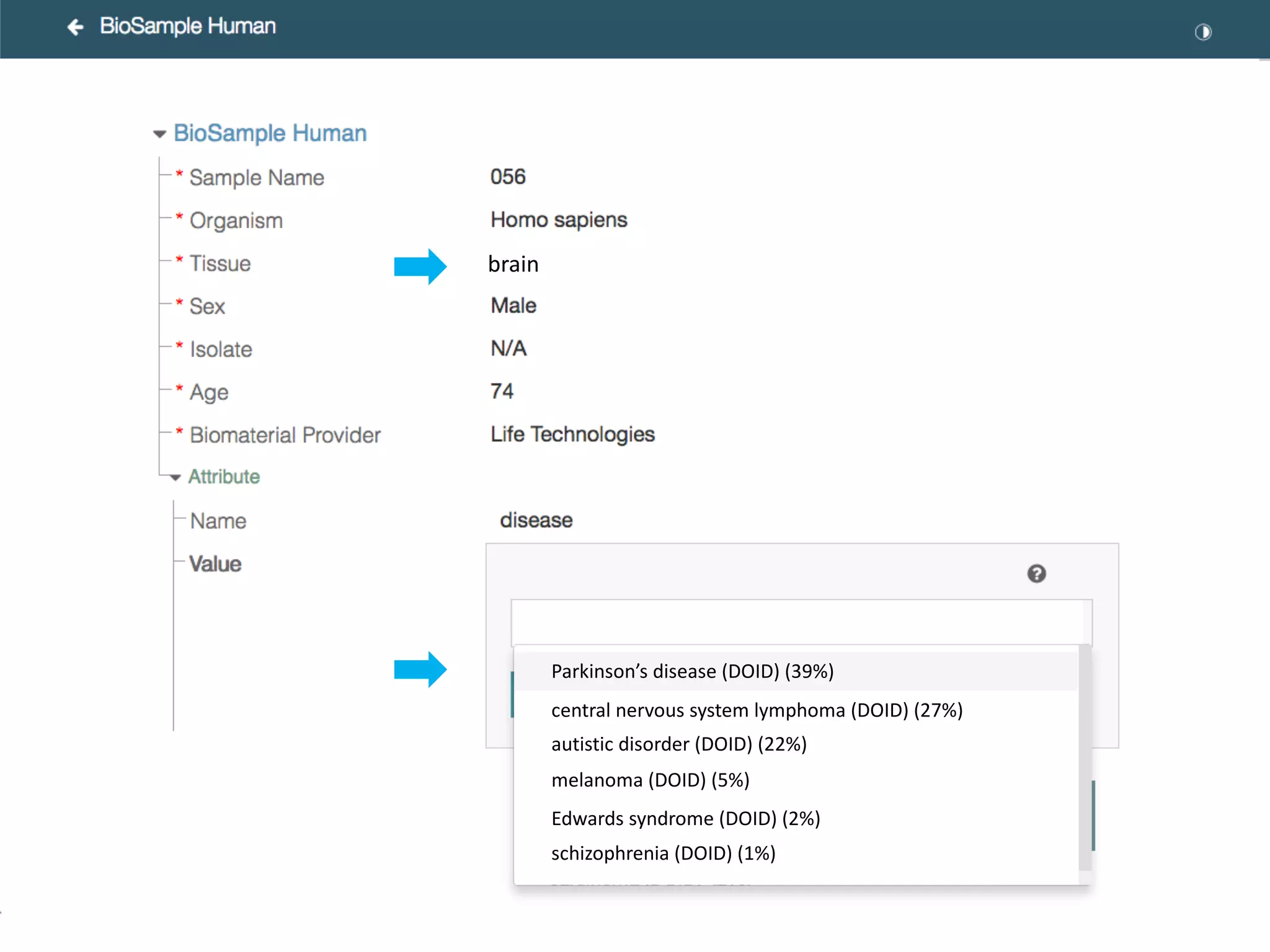
The document discusses the challenges and solutions in metadata management for scientific data sharing, highlighting the importance of accessible, clearly defined, and consistently annotated datasets. Purvesh Khatri's work exemplifies how 'data parasites' can derive significant insights from existing data without direct experimental work, emphasizing the need for standardized metadata and open access. The Cedar Workbench, a proposed solution, aims to create a structured ecosystem for generating and managing experimental metadata, thereby improving data usability and fostering a culture of data sharing in the scientific community.









![age
Age
AGE
`Age
age (after birth)
age (in years)
age (y)
age (year)
age (years)
Age (years)
Age (Years)
age (yr)
age (yr-old)
age (yrs)
Age (yrs)
age [y]
age [year]
age [years]
age in years
age of patient
Age of patient
age of subjects
age(years)
Age(years)
Age(yrs.)
Age, year
age, years
age, yrs
age.year
age_years
Failure to use standard terms makes
datasets often impossible to search](https://image.slidesharecdn.com/fairomcoiwebinarcedarworkbenchformetadatamanagementmarkmusen12dec2019-191216140526/75/CEDAR-work-bench-for-metadata-management-10-2048.jpg)






























![age
Age
AGE
`Age
age (after birth)
age (in years)
age (y)
age (year)
age (years)
Age (years)
Age (Years)
age (yr)
age (yr-old)
age (yrs)
Age (yrs)
age [y]
age [year]
age [years]
age in years
age of patient
Age of patient
age of subjects
age(years)
Age(years)
Age(yrs.)
Age, year
age, years
age, yrs
age.year
age_years](https://image.slidesharecdn.com/fairomcoiwebinarcedarworkbenchformetadatamanagementmarkmusen12dec2019-191216140526/75/CEDAR-work-bench-for-metadata-management-50-2048.jpg)