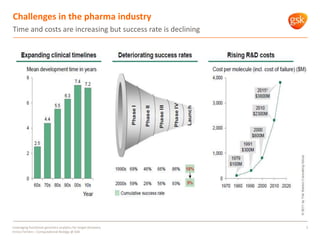
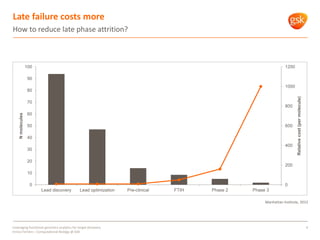
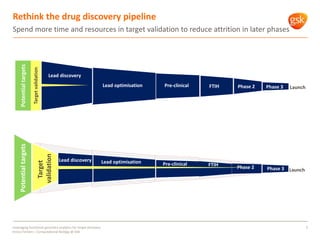
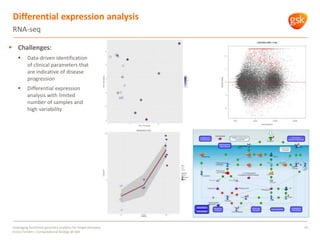
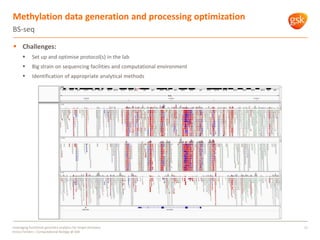
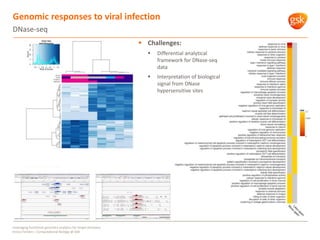
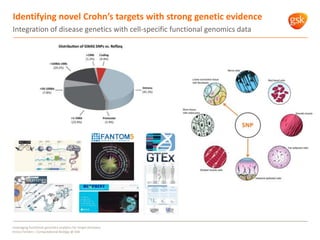
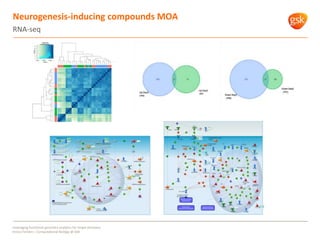
The document discusses how GSK is leveraging functional genomics analytics like RNA-seq, ChIP-seq, and DNase-seq to improve target discovery. It provides examples of projects analyzing disease progression in rheumatoid arthritis, responses to viral infection, and mechanisms of action of neurogenesis compounds. The goal is to better understand diseases, targets, and drugs to reduce late stage clinical trial failures and costs through more rigorous target validation early on. Partnerships with academic institutions are also highlighted.