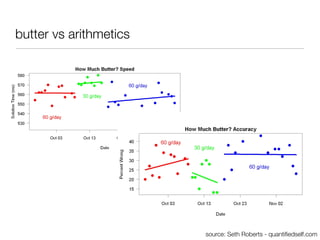
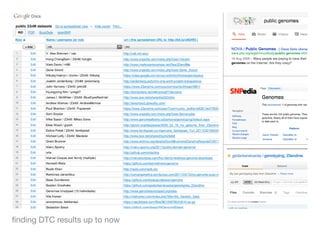
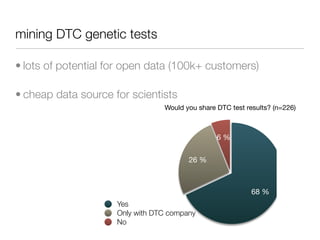
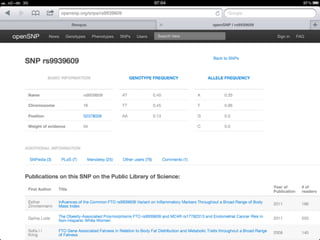
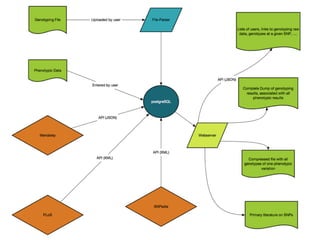
The document discusses the potential for using direct-to-consumer (DTC) genetic testing results and self-tracked health data from the Quantified Self movement to enable new types of genome-wide association studies and medical research. It outlines plans to create an open-source platform called openSNP that would allow users to voluntarily share their DTC genetic test results and phenotypes. This could generate large datasets for scientific studies while lowering costs compared to traditional research methods. However, challenges include ensuring representative sample sizes and accurate self-reported user data. Positive examples from other platforms like 23andMe and PatientsLikeMe that have successfully conducted research using crowdsourced data are provided.










![positive examples: 23andMe – general traits
“
Replications of associations [...] for hair color, eye color,
and freckling validate the Web-based, self-reporting
paradigm. The identification of novel associations for hair
morphology [...], freckling [...], the ability to smell the
methanethiol produced after eating asparagus [...], and
photic sneeze reflex [...] illustrates the power of the
approach.
Nicolas Eriksson et al. (2010) Web-Based, Participant-Driven Studies Yield Novel
Genetic Associations for Common Traits. PLoS Genet 6(6): e1000993. doi:10.1371/
journal.pgen.1000993](https://image.slidesharecdn.com/tuebingen-120216051428-phpapp02/85/openSNP-Crowdsourcing-Genome-Wide-Association-Studies-21-320.jpg)
![positive examples: 23andMe – Parkinson’s Disease
“
We discovered two novel, genome-wide significant
associations with [Parkinson’s Disease]—both replicated
in an independent cohort. We also replicated 20
previously discovered genetic associations (including
LRRK2, GBA, SNCA, MAPT, GAK, and the HLA region),
providing support for our novel study design.
Chuong B. Do et al. (2011) Web-Based Genome-Wide Association Study Identifies
Two Novel Loci and a Substantial Genetic Component for Parkinson's Disease. PLoS
Genet 7(6): e1002141. doi:10.1371/journal.pgen.1002141](https://image.slidesharecdn.com/tuebingen-120216051428-phpapp02/85/openSNP-Crowdsourcing-Genome-Wide-Association-Studies-22-320.jpg)