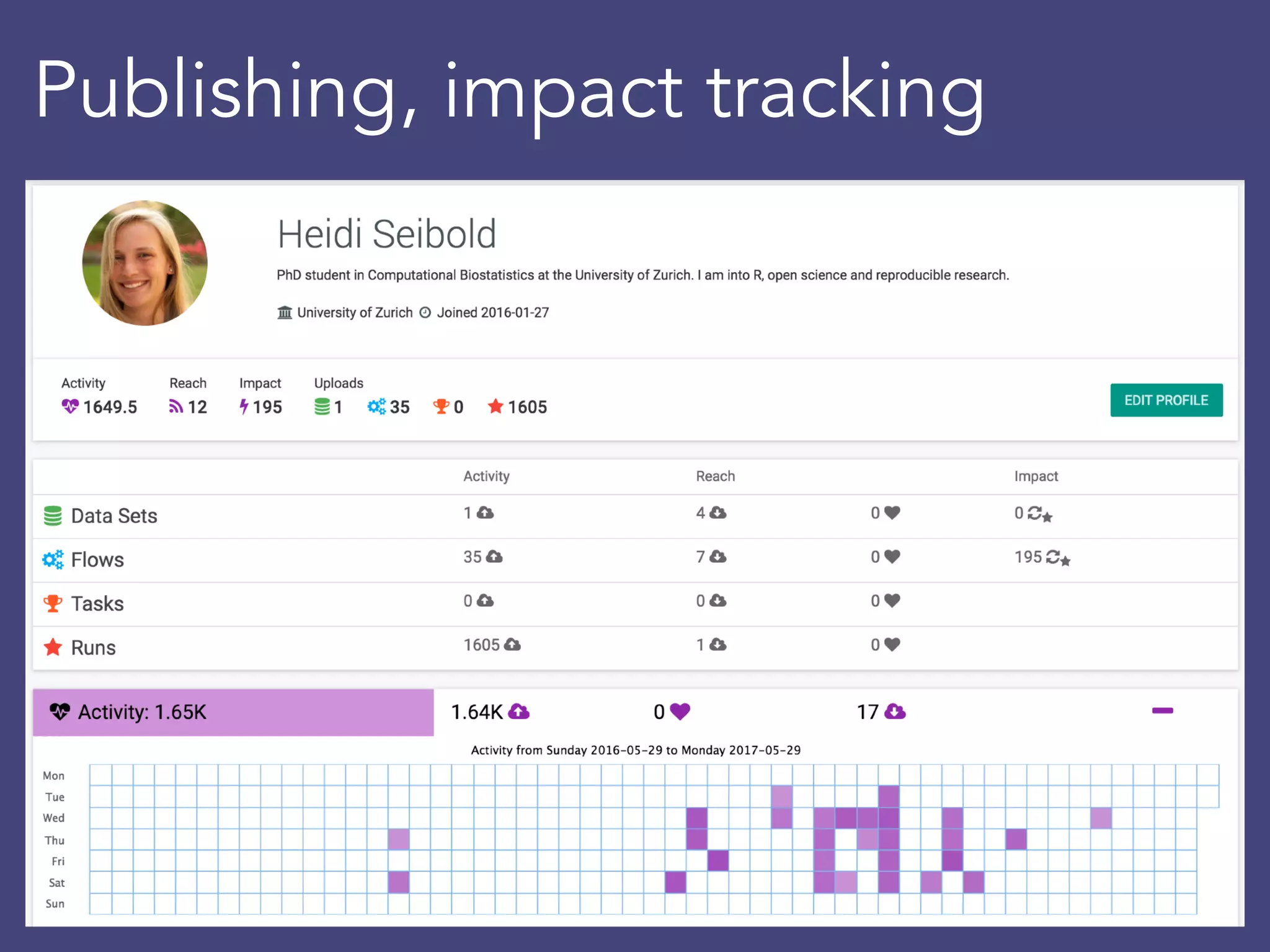
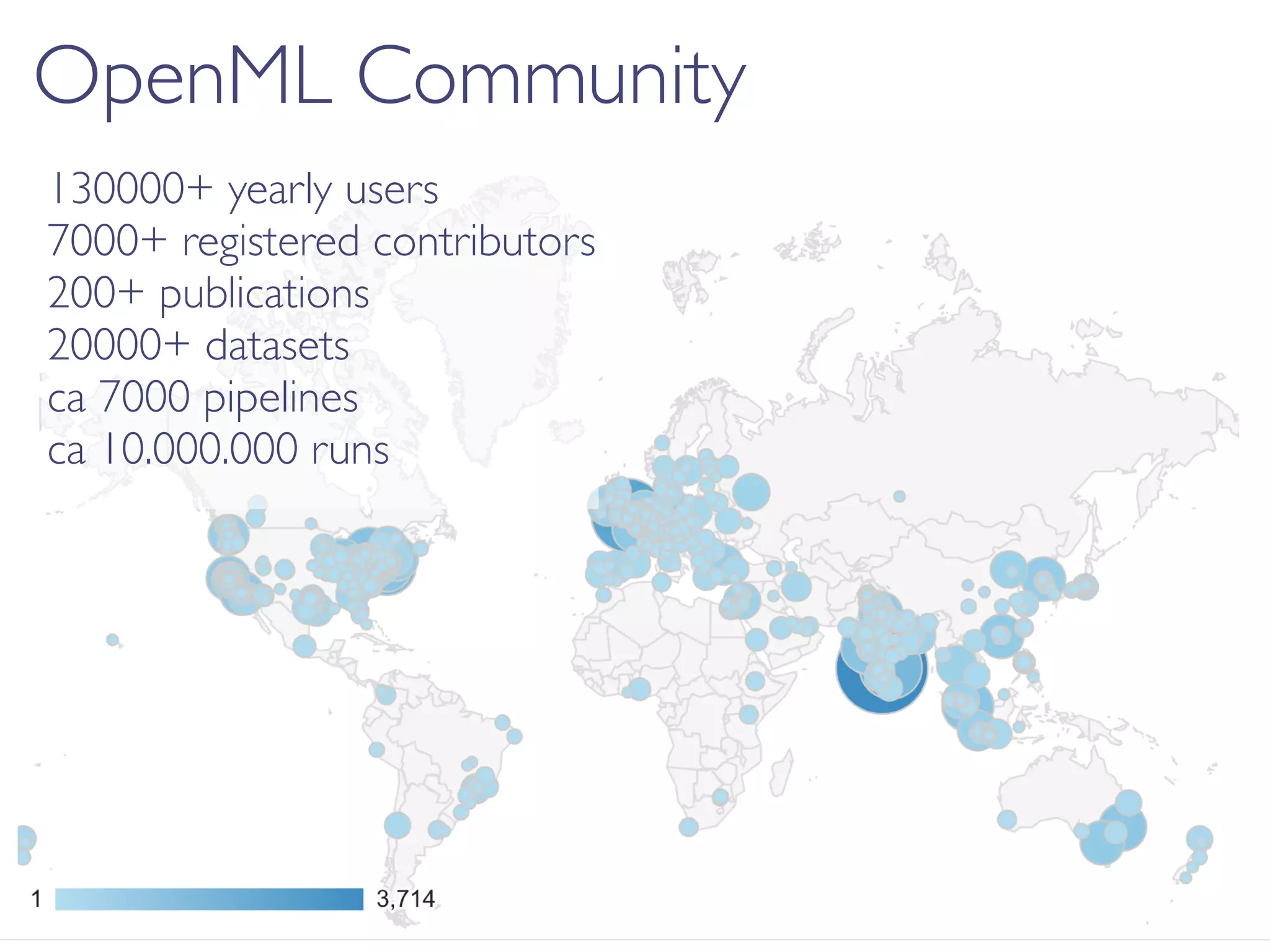
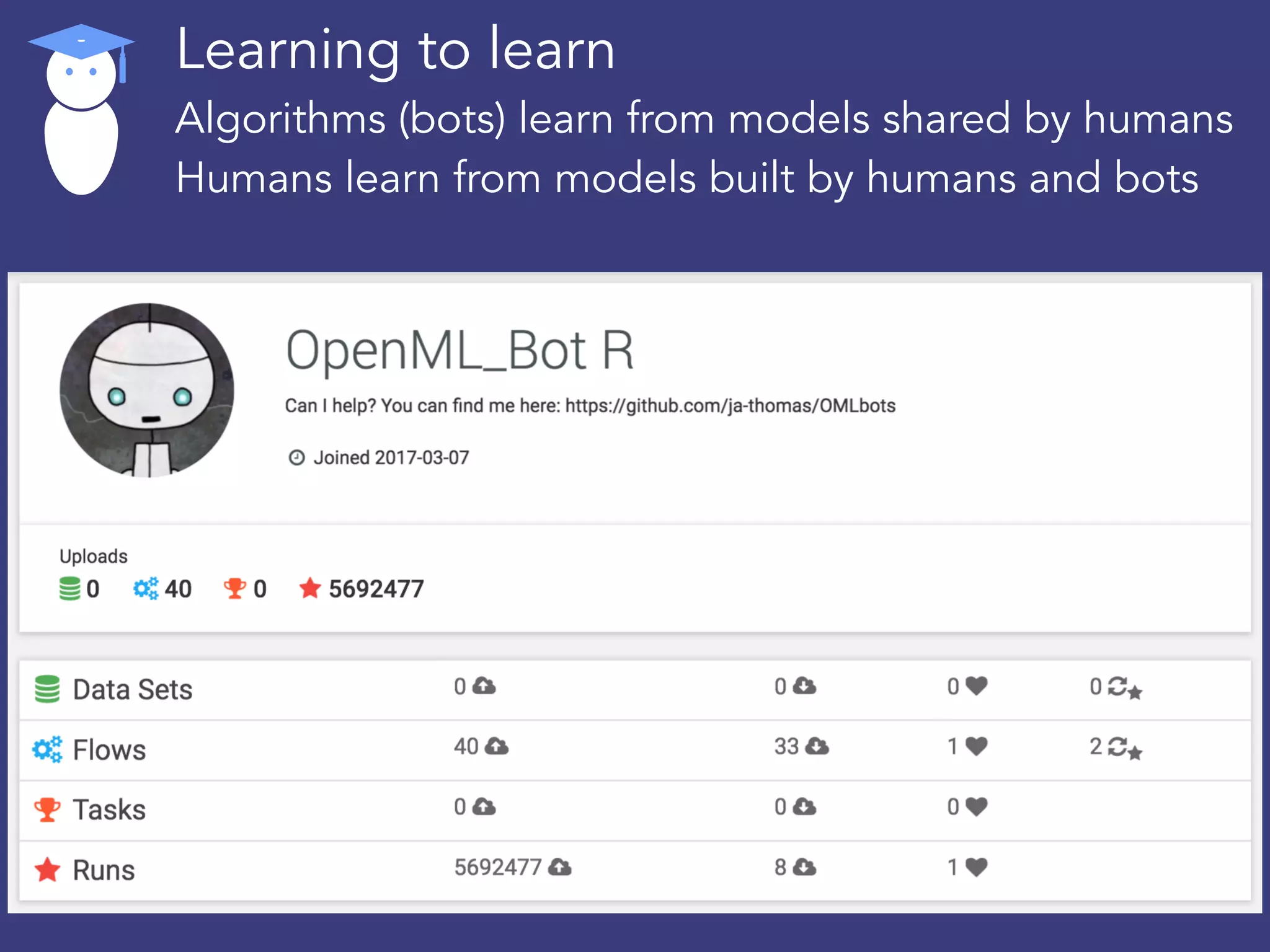
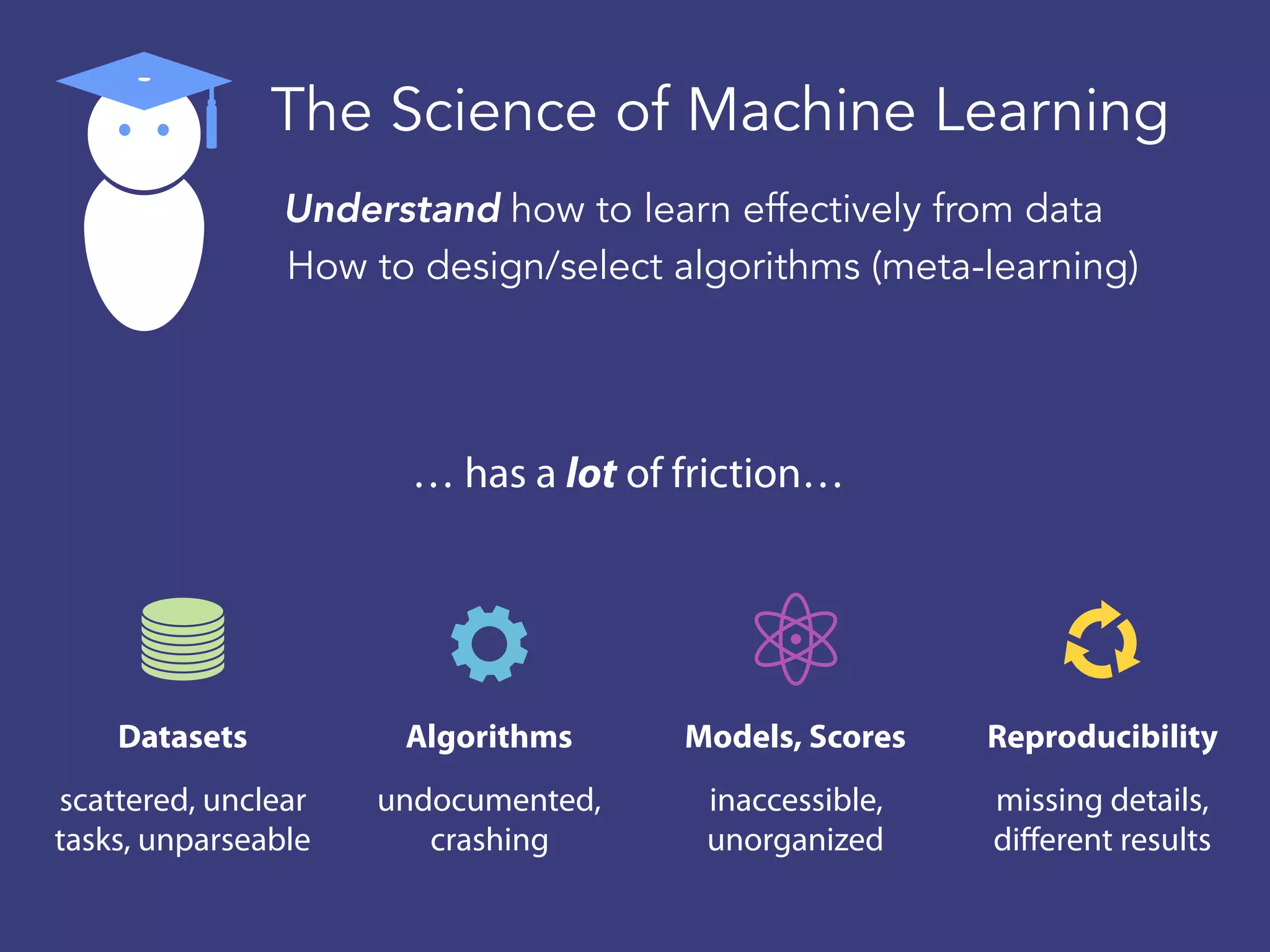
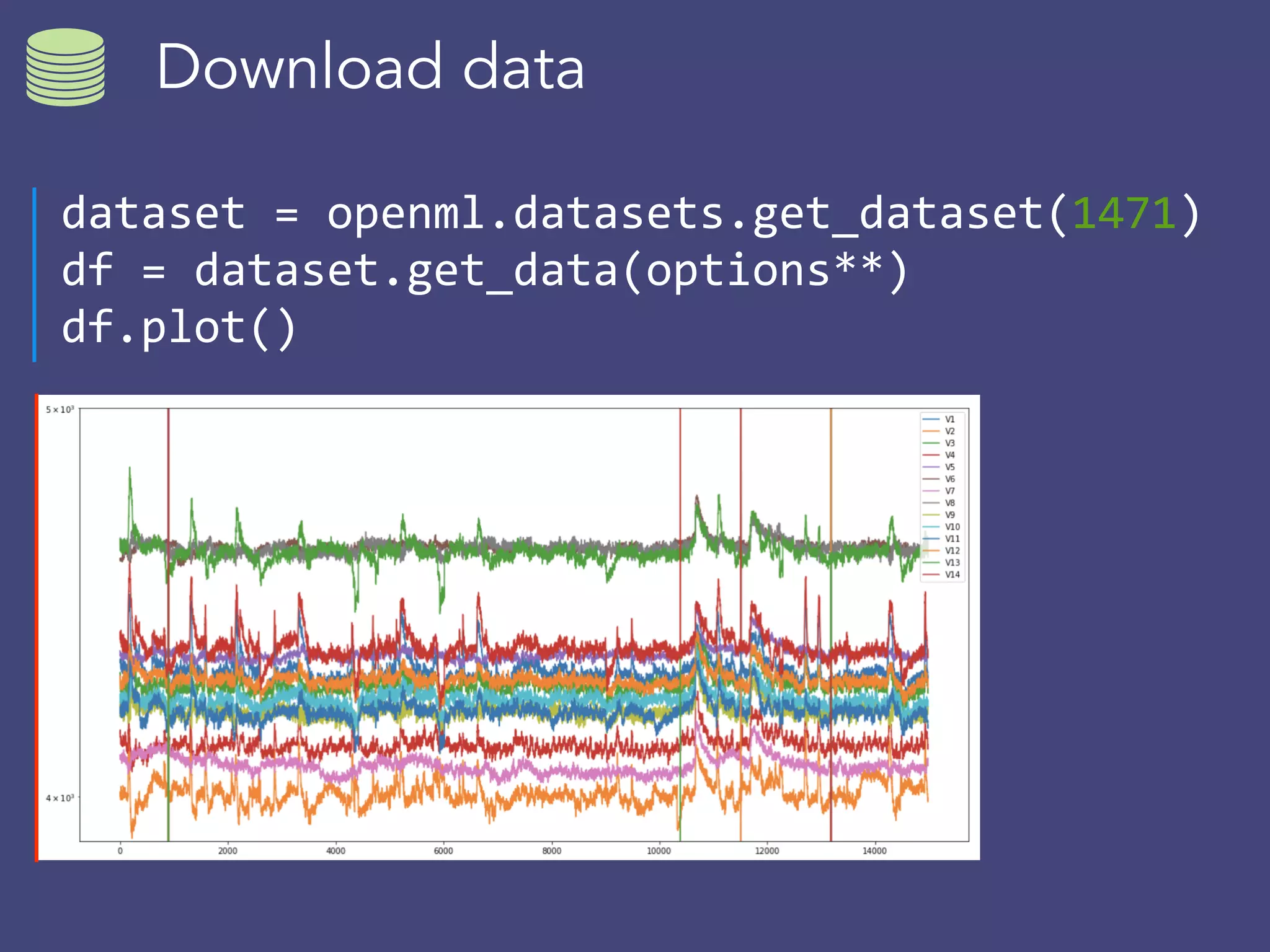
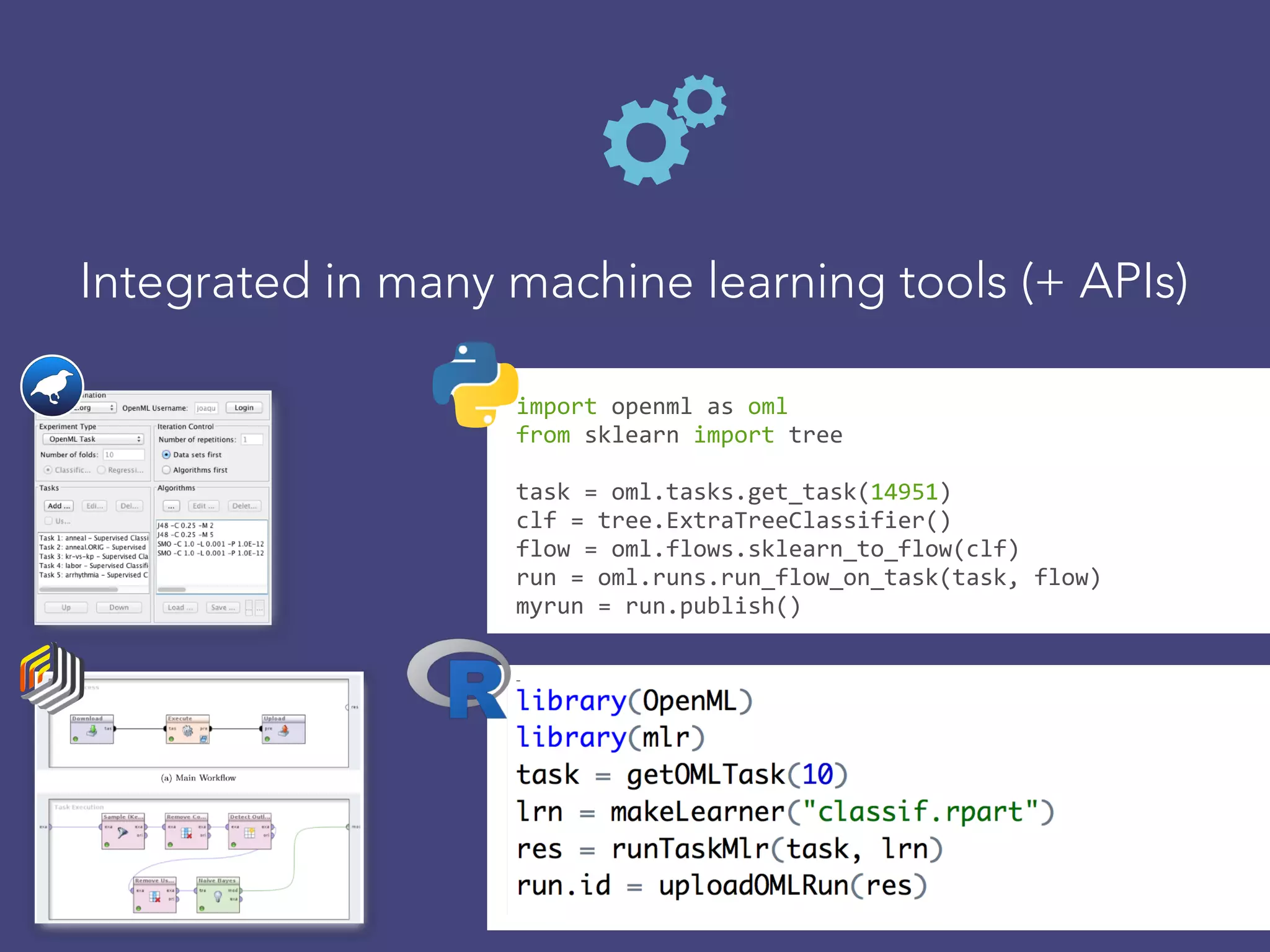
OpenML is a collaborative platform aimed at improving machine learning by organizing datasets, algorithms, and models for easier access and reproducibility. It provides tools to share data, integrate workflows across various programming languages, and enables users to automatically generate metadata for seamless discovery and evaluation. With a vibrant community, OpenML fosters enhanced collaboration and innovation in the machine learning field through automated learning and benchmarking capabilities.
























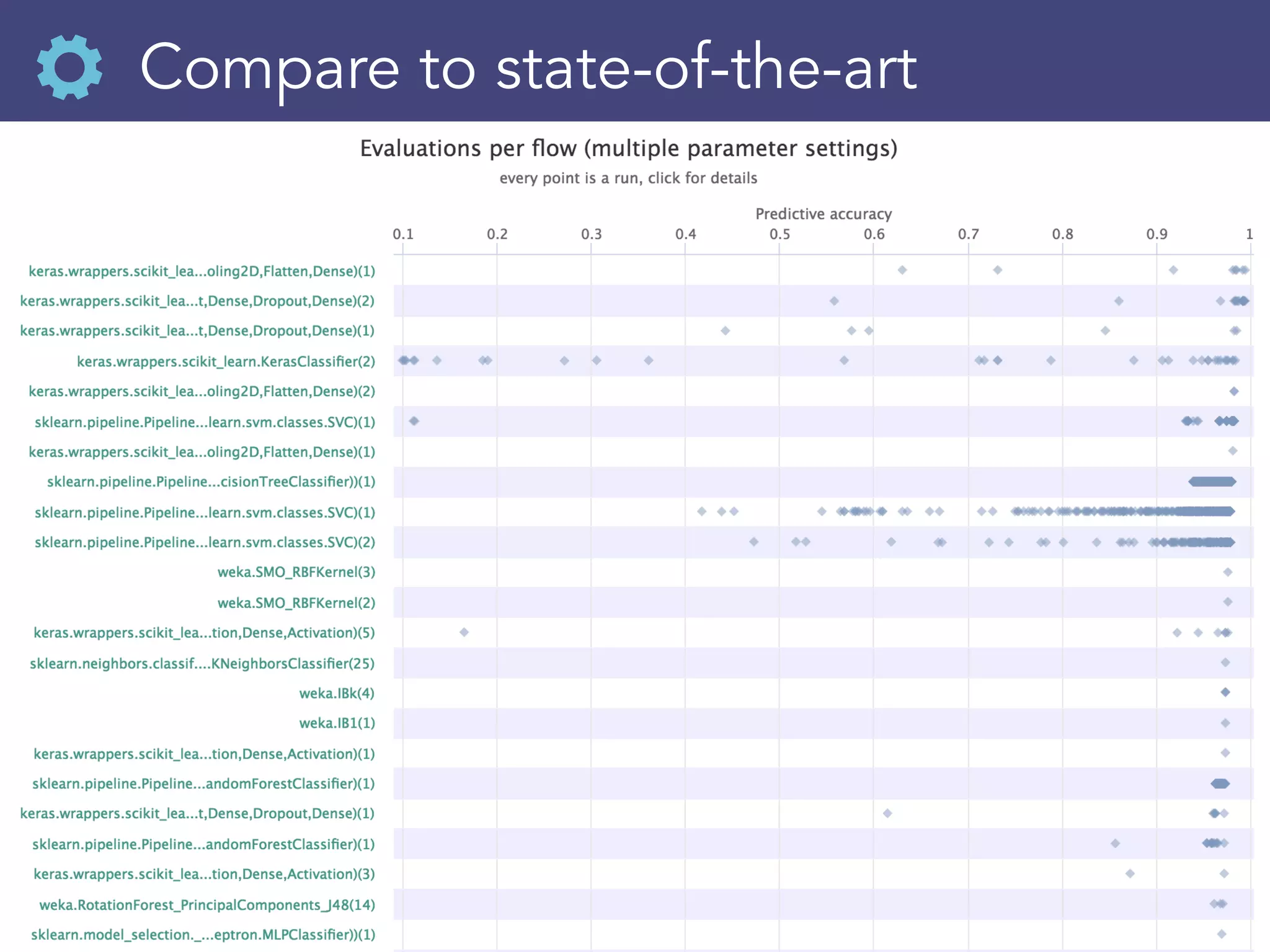


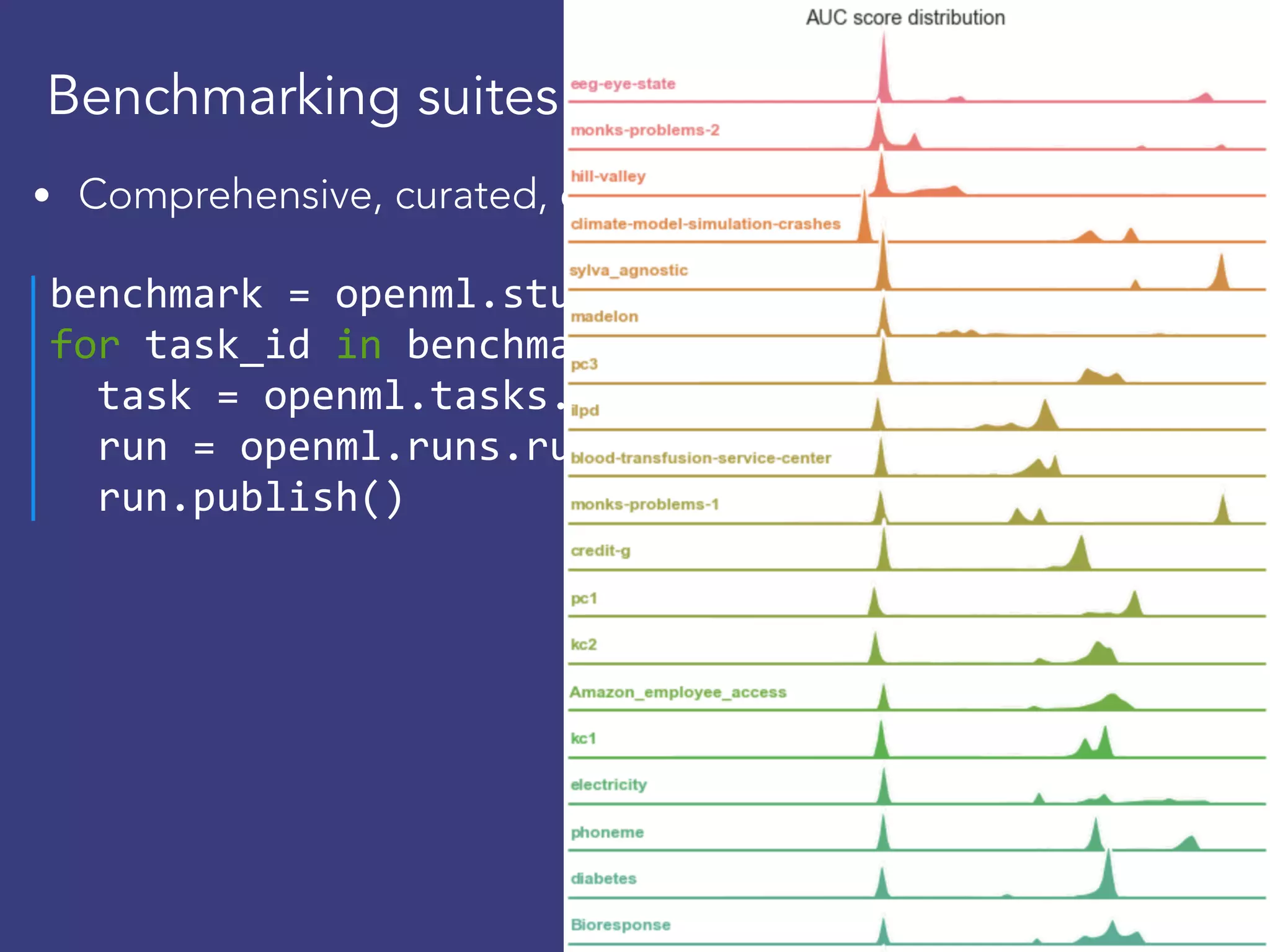
![Download, reuse runs
evals = openml.evaluations.list_evaluations(
task=[145677], function=‘area_under_roc_curve’)
scores =[{"flow":e.flow_name, "score":e.value} for id,
sns.violinplot(x="score", y="flow", data= pd.DataFrame](https://image.slidesharecdn.com/openml2019-190325235230/75/OpenML-2019-32-2048.jpg)