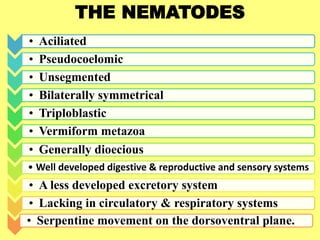
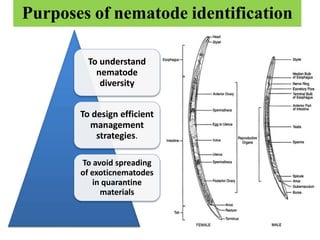
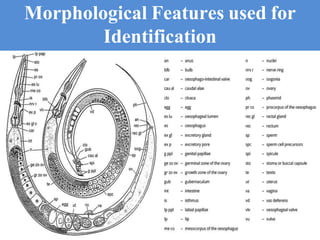
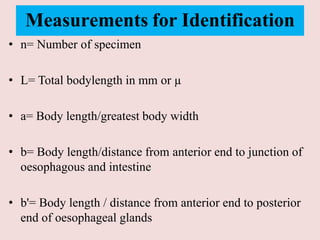
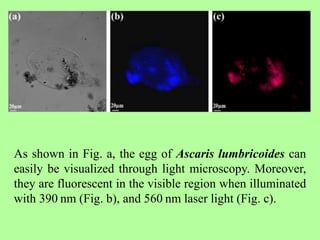
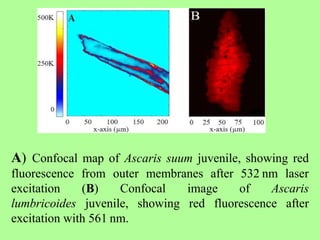
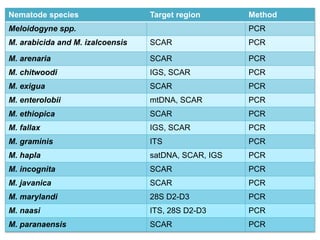
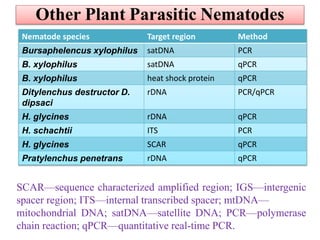
The document discusses recent advancements in nematode identification techniques, emphasizing the importance of accurate identification for effective management strategies. It covers various morphological, biochemical, and molecular methods, including machine learning, autofluorescence, isozymes, and DNA-based analysis, as well as their advantages and limitations. The conclusion highlights the dependence on molecular methods for distinguishing nematodes at the species and subspecies levels to enable focused management approaches.