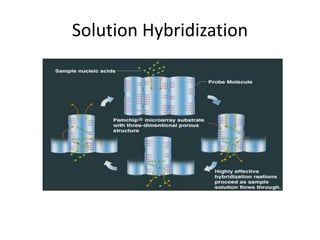
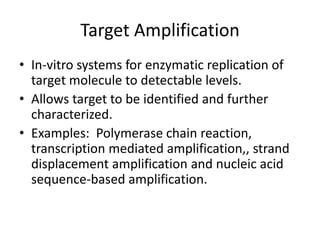
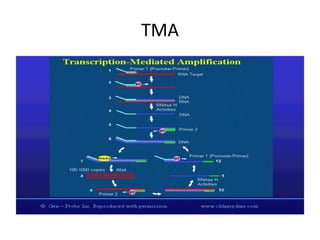
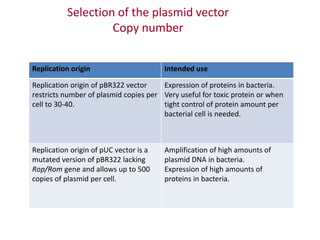
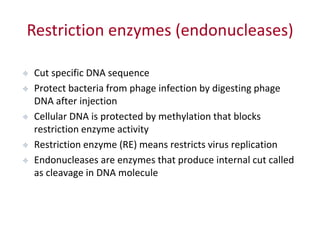
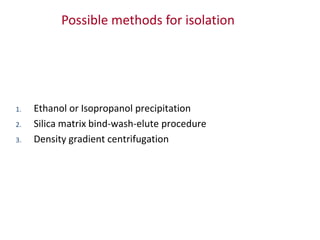
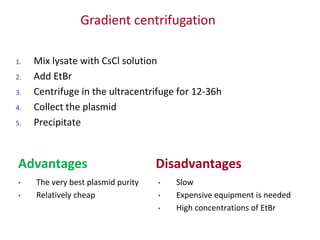
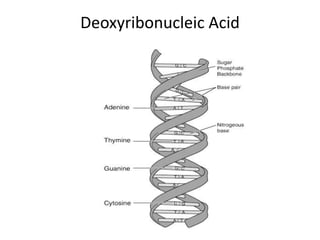
Plasmids are small, extra-chromosomal DNA molecules capable of replicating independently of chromosomal DNA. They are commonly used as vectors to introduce foreign DNA into host cells. Restriction enzymes cut DNA at specific recognition sequences and are used in molecular cloning. Various techniques like gel electrophoresis, Southern blot, and DNA microarrays can analyze DNA sequences.
































![Southern Blot
• MOM [blue], DAD [yellow], and their four children: D1 (the biological
daughter), D2 (step-daughter, child of Mom and her former husband
[red]), S1 (biological son), and S2 (adopted son,not biologically related [his
parents are light and dark green]).](https://image.slidesharecdn.com/moleculartechniques-170220141025/85/Molecular-techniques-38-320.jpg)