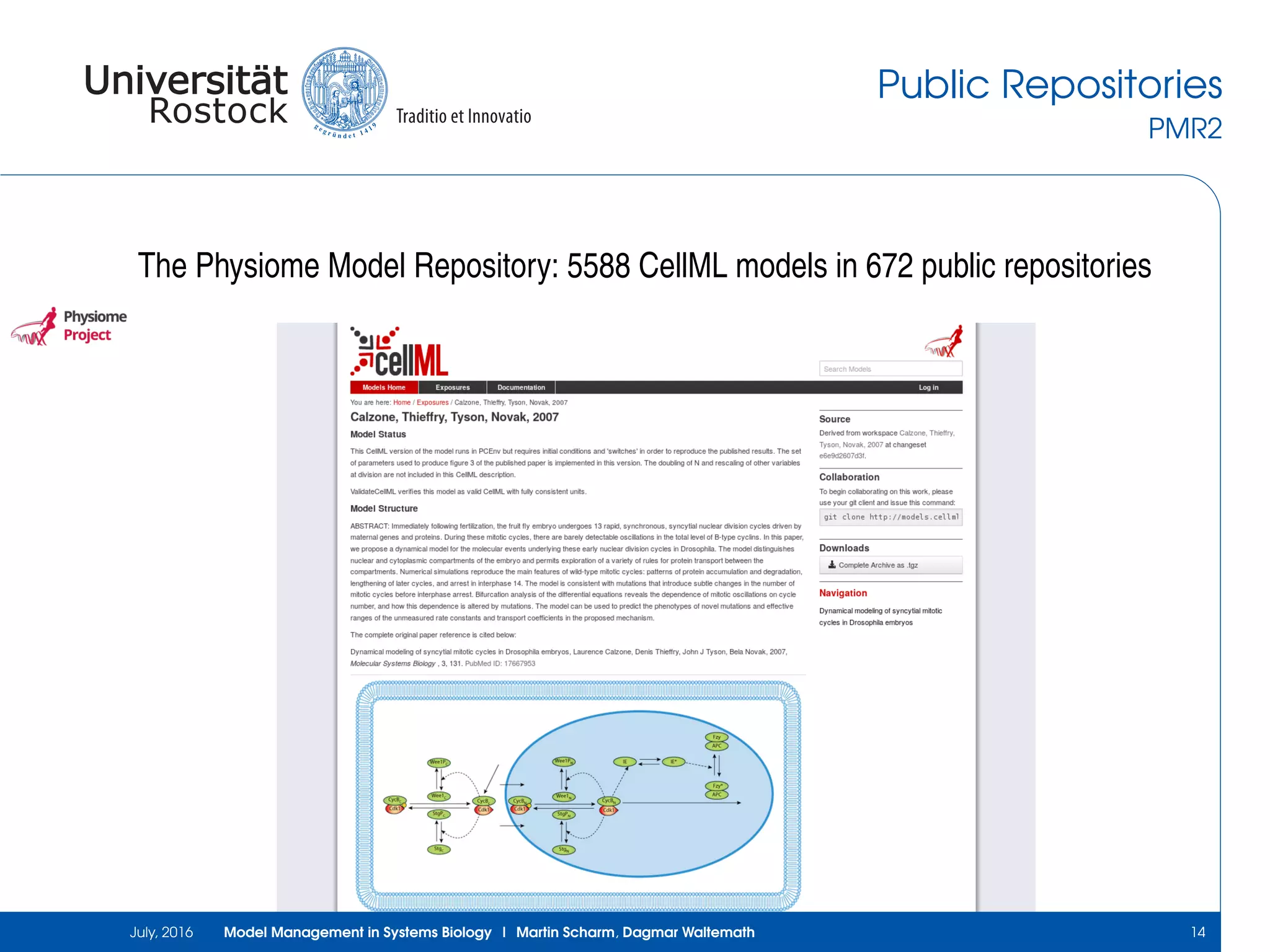
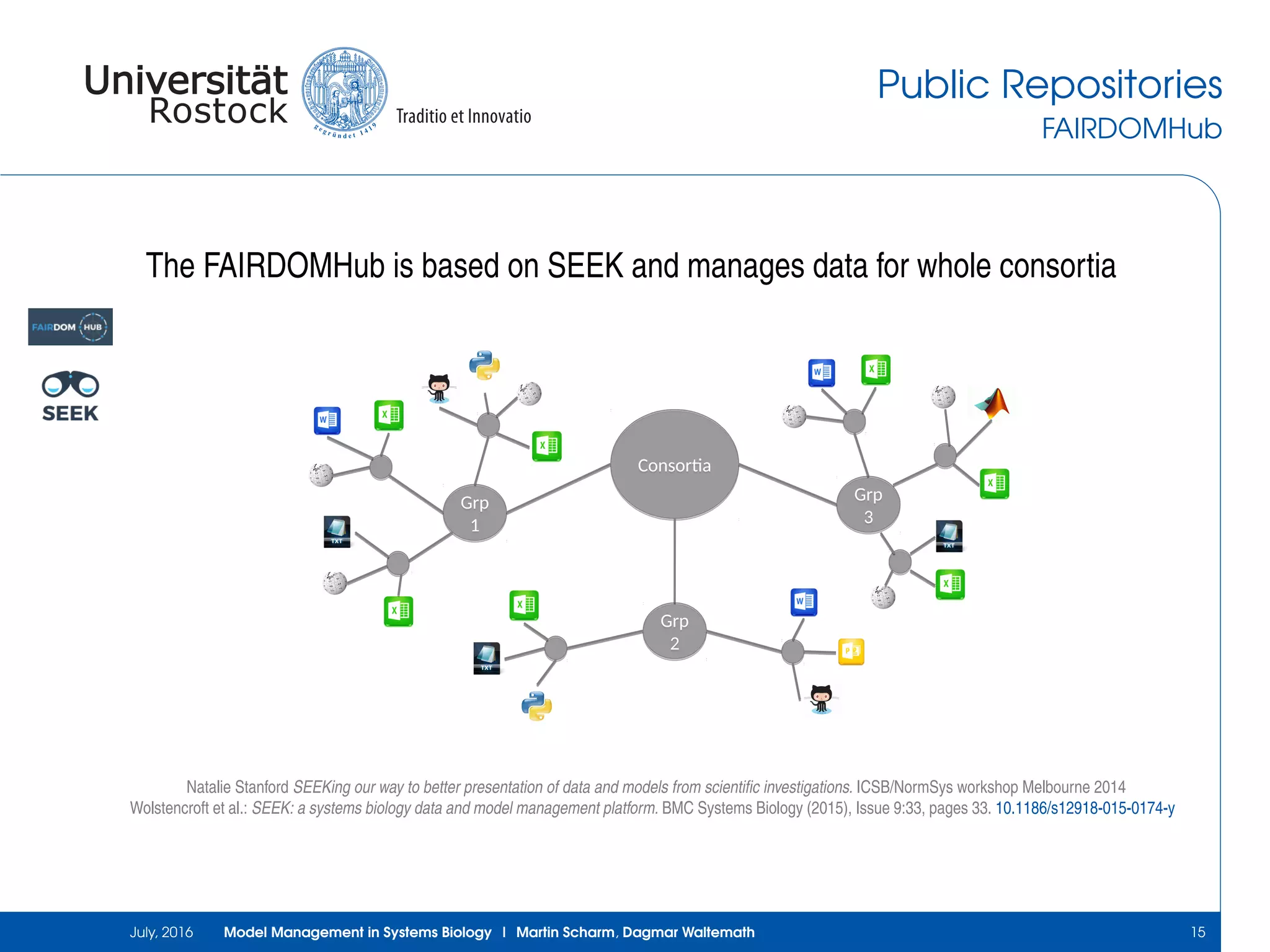
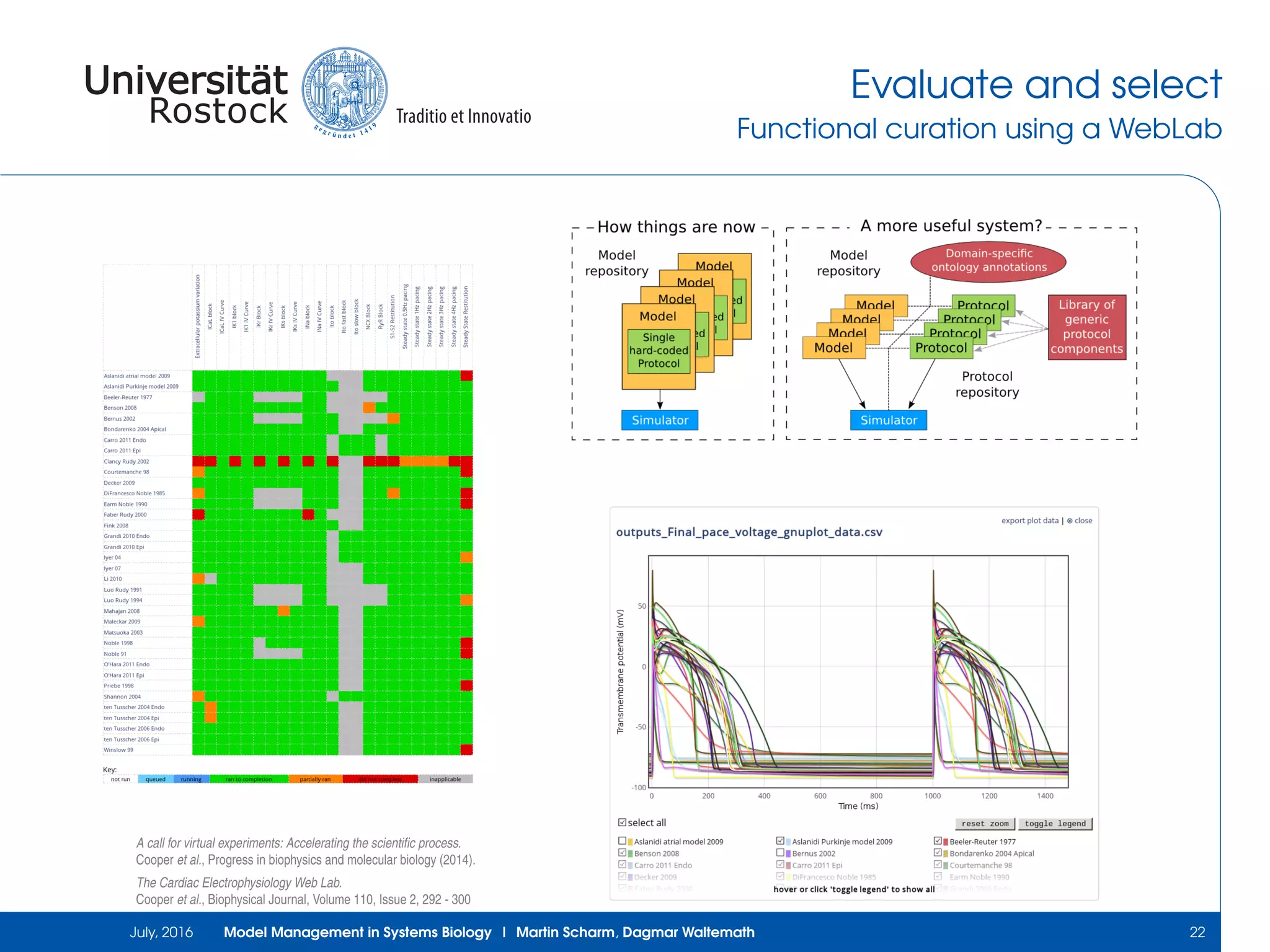
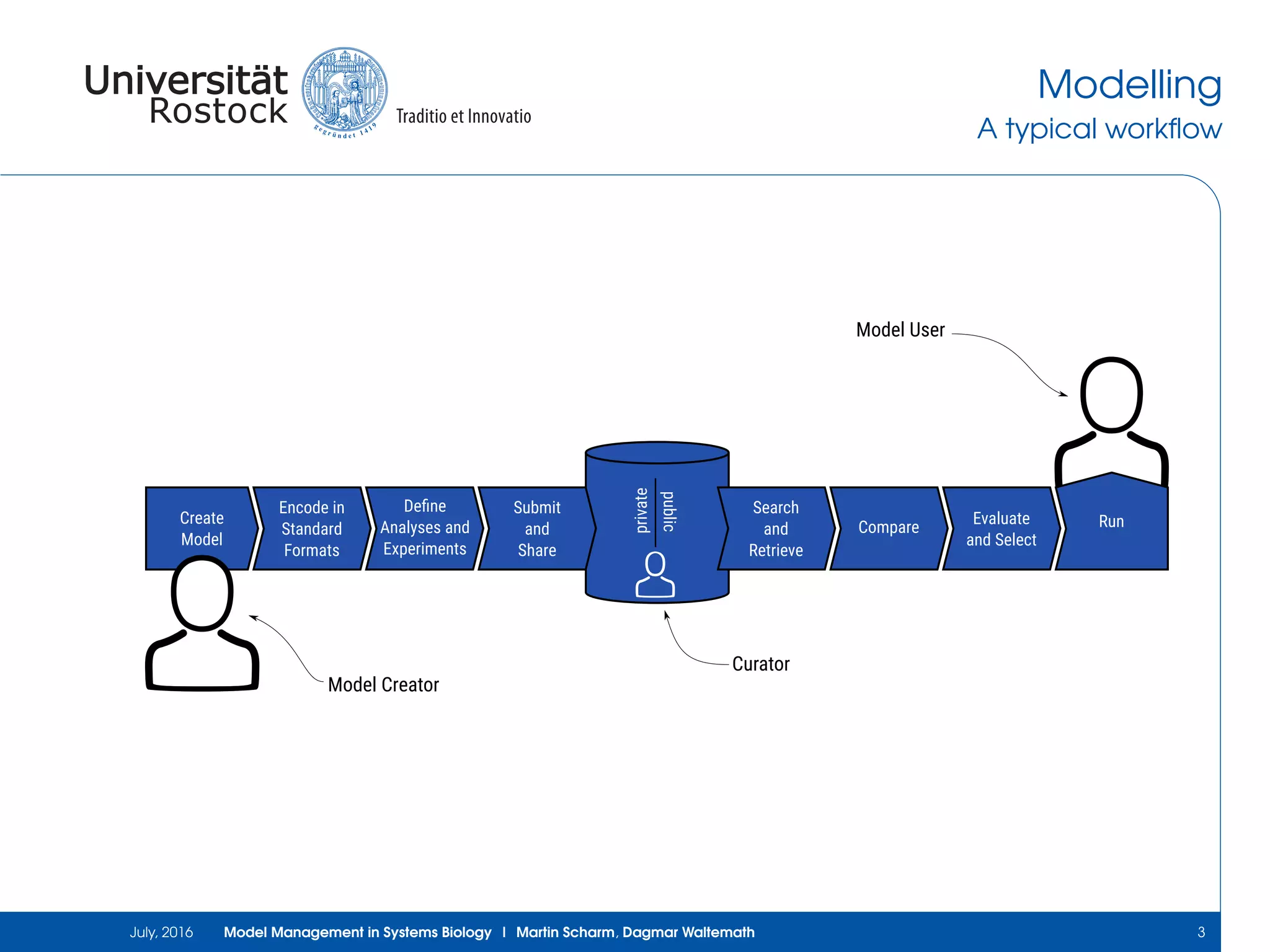
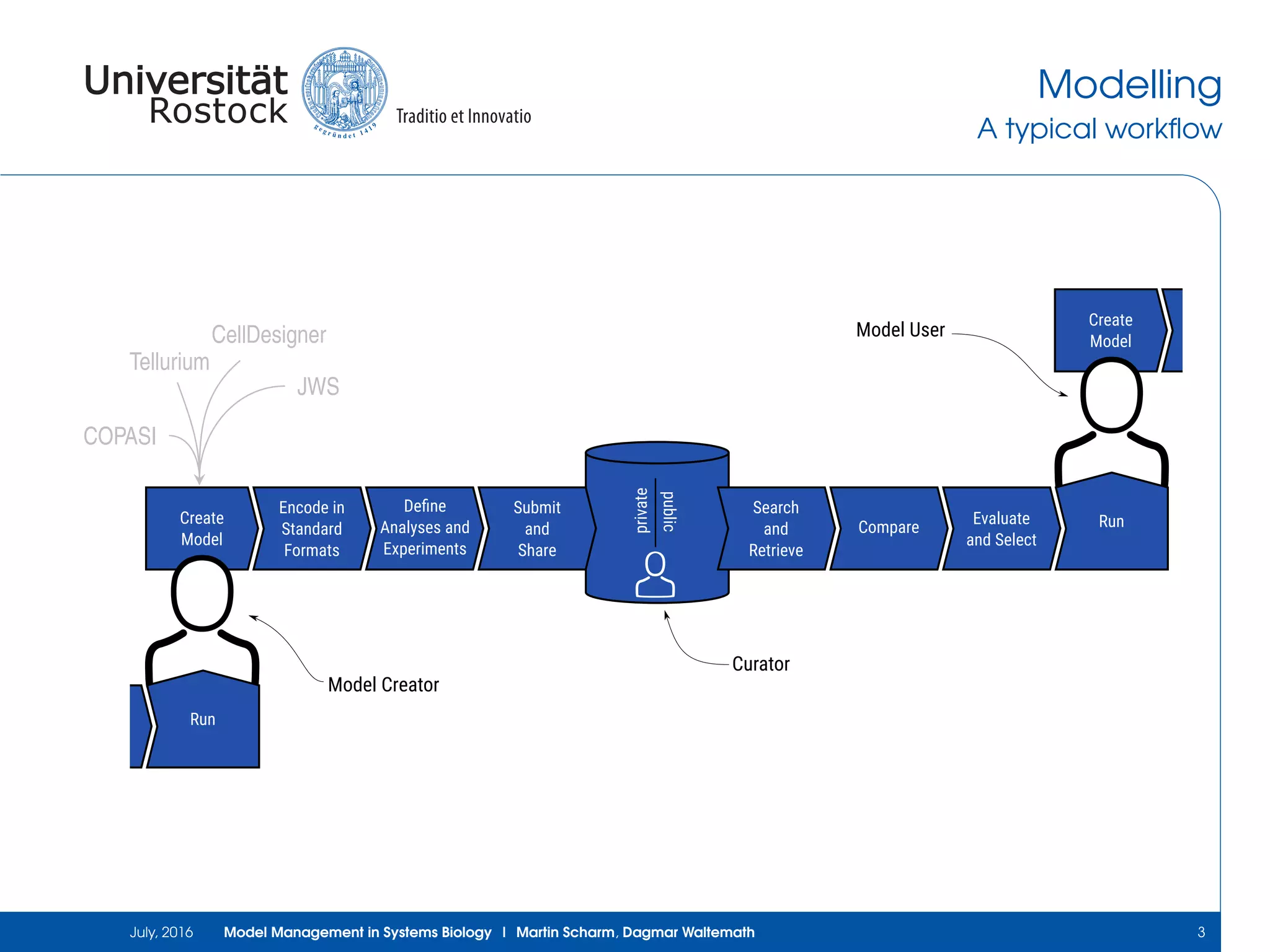
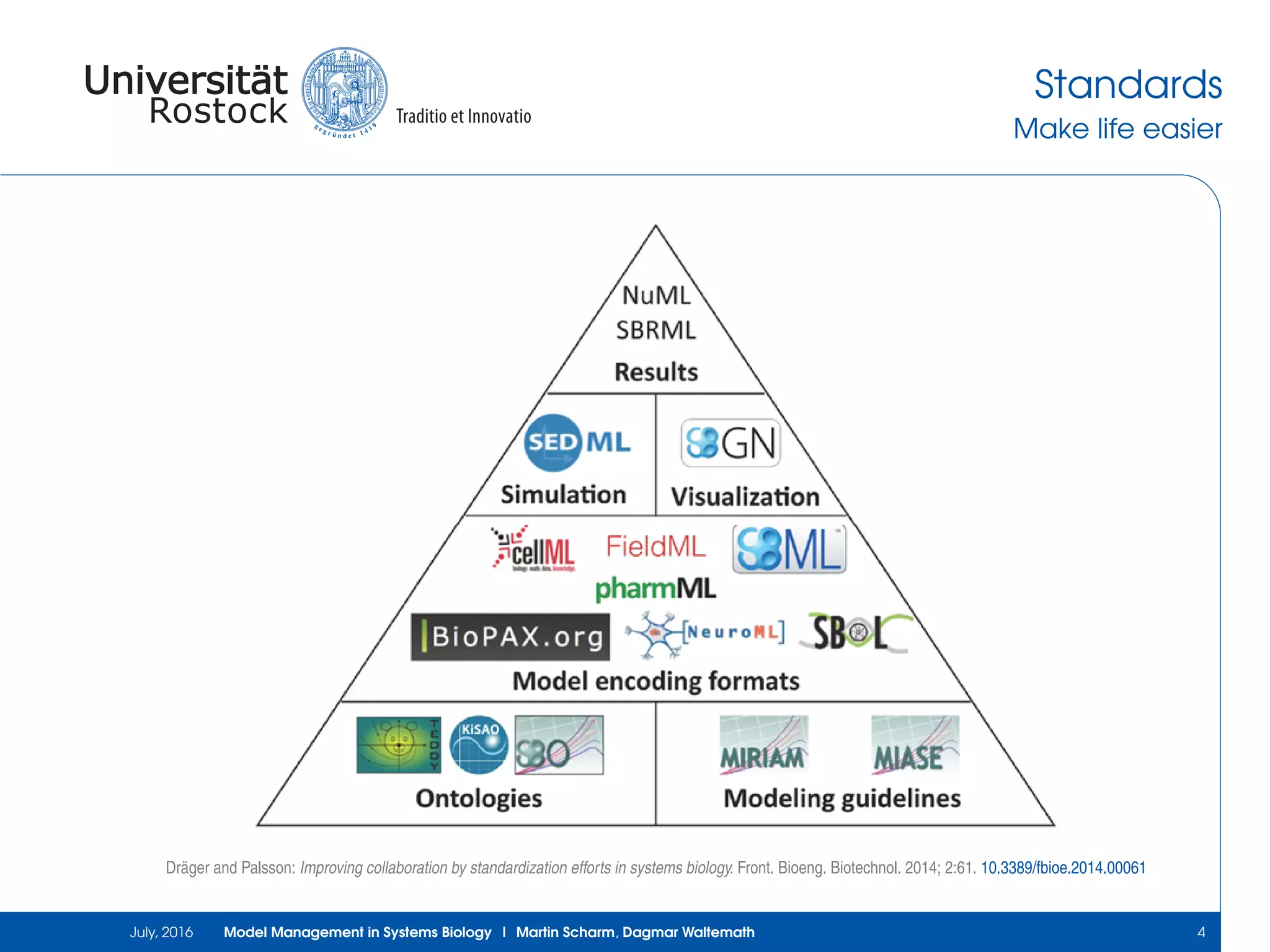
The document discusses the challenges and approaches in model management within systems biology and bioinformatics, emphasizing the increasing complexity and number of models. It outlines workflows for modeling, sharing research, and the importance of standards for collaboration, while also highlighting ongoing challenges such as version control and data provenance. The authors, Martin Scharm and Dagmar Waltemath, aim to improve accessibility and reproducibility of simulation studies in the field.
![Generate an Experiment
Encoding the simulation study
Calzone et al. (2007): Dynamical modeling of syncytial mitotic
cycles in Drosophila embryos. Mol Syst Biol. 3: 131
TM
Wee1n_1 MPFn_1 StgPn_1
mol/l
0
0.2
0.4
0.6
0.8
1
1.2
1.4
s
0 50 100 150 200 250
MPFn_1 StgPn_1 Wee1n_1
mol/l
0
0.2
0.4
0.6
0.8
1
1.2
s
0 50 100 150 200 250
[MPFc]|Time [MPFn]|Time [Stgc]|Time [Stgn]|Time [Wee1n]|Time [preMPFc]|Time [preMPFn]|Time
mol/l
0
0.5
1
1.5
2
s
0 50 100 150 200 250
MLSED
as published MLSED
modified initial
environment
MLSED
selected different species
adapted from Waltemath: Reproducible virtual experiments with SED-ML. Harmony 2016, Auckland, NZ
July, 2016 Model Management in Systems Biology | Martin Scharm, Dagmar Waltemath 5](https://image.slidesharecdn.com/fairdom-webinar-model-management-scharm-160726132711/75/Model-Management-in-Systems-Biology-Challenges-Approaches-Solutions-6-2048.jpg)