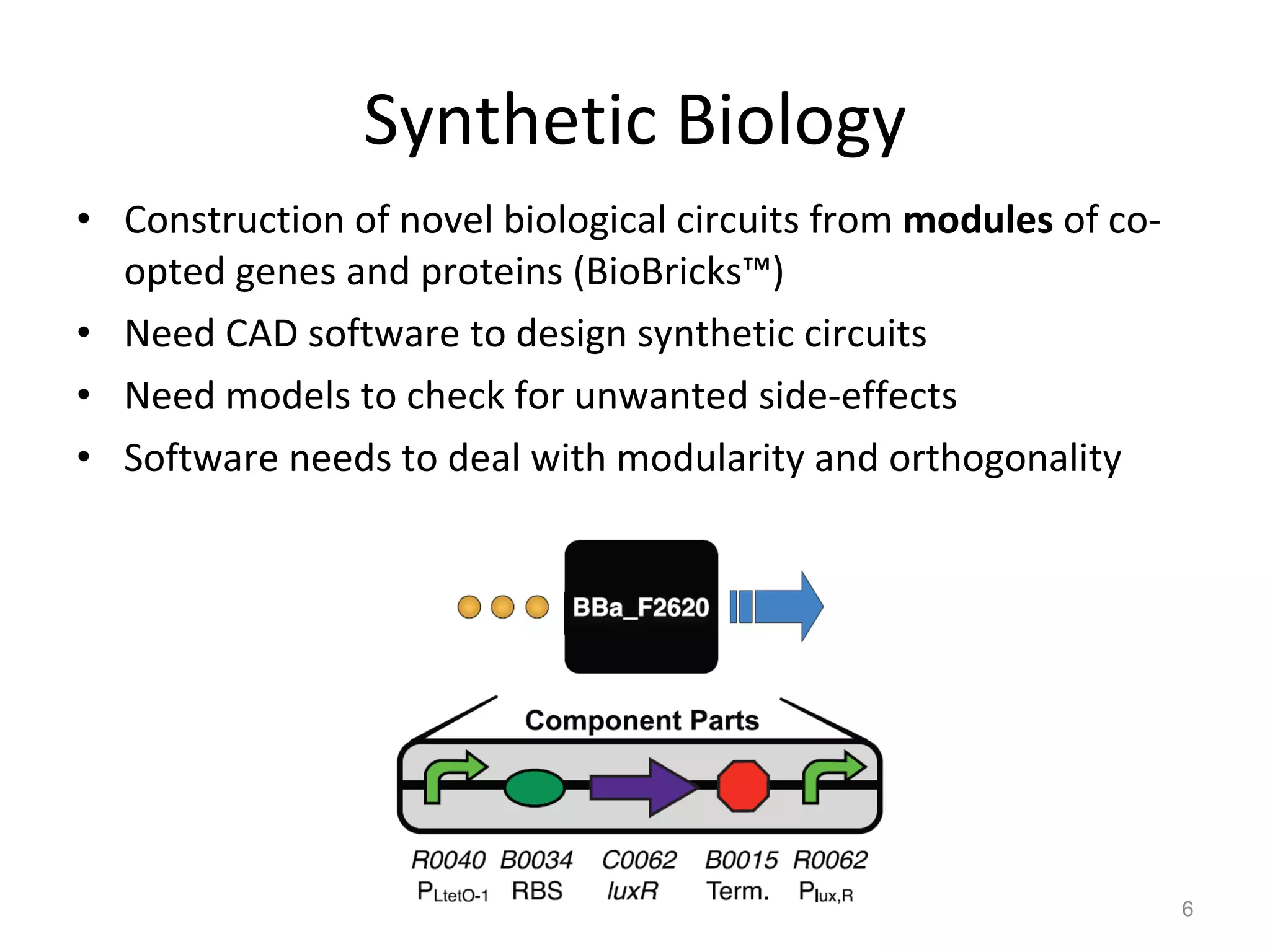
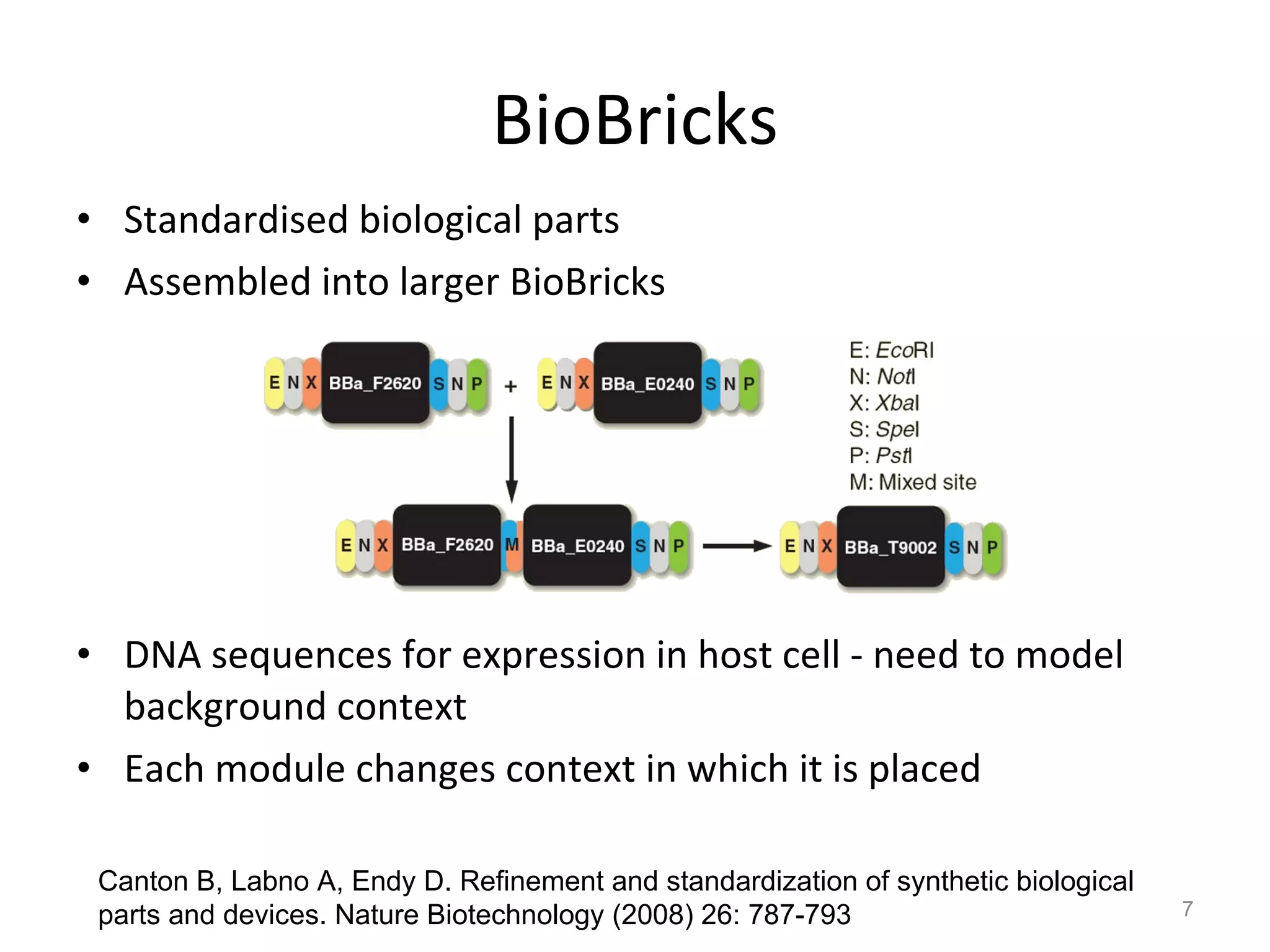
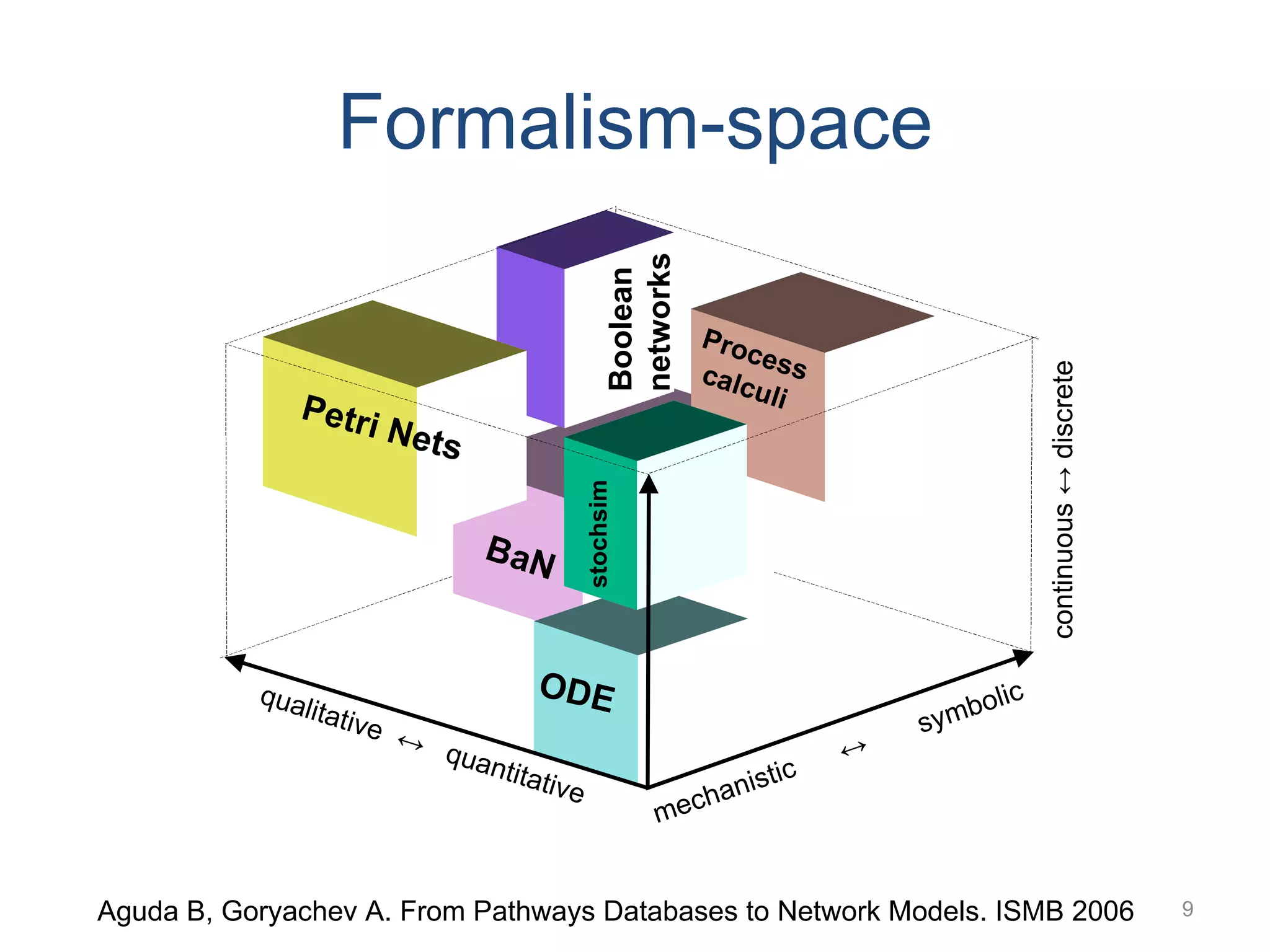
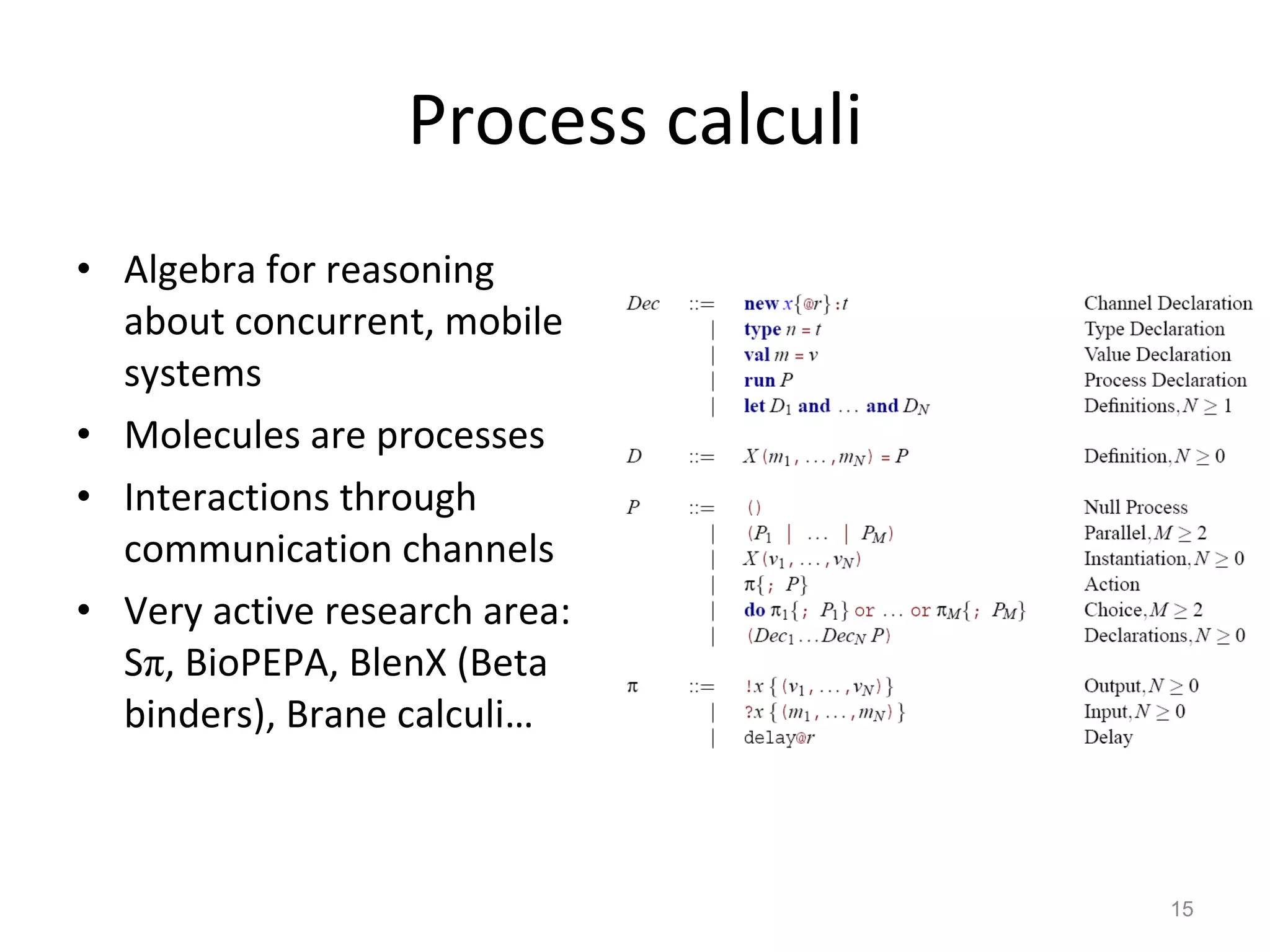
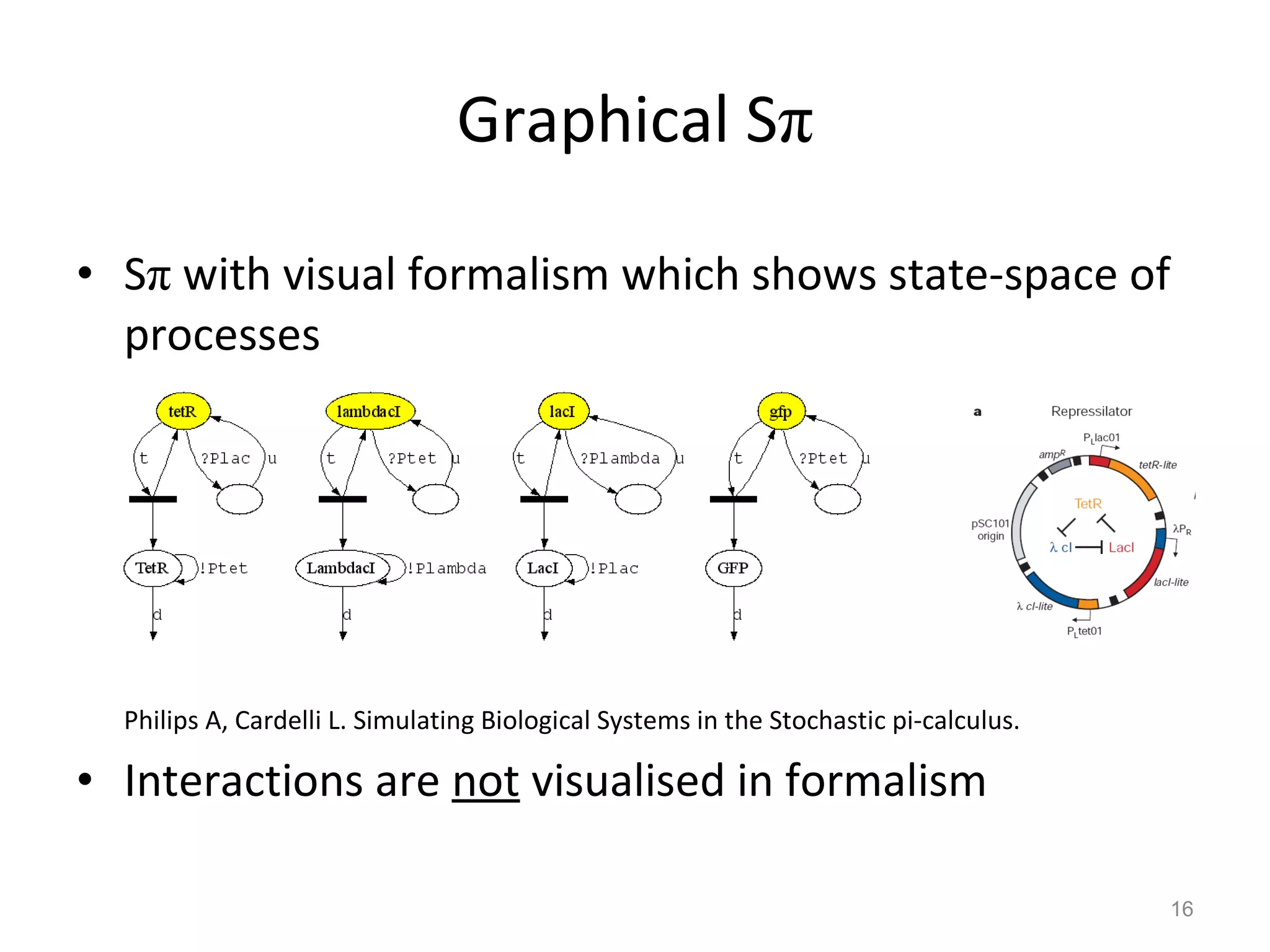
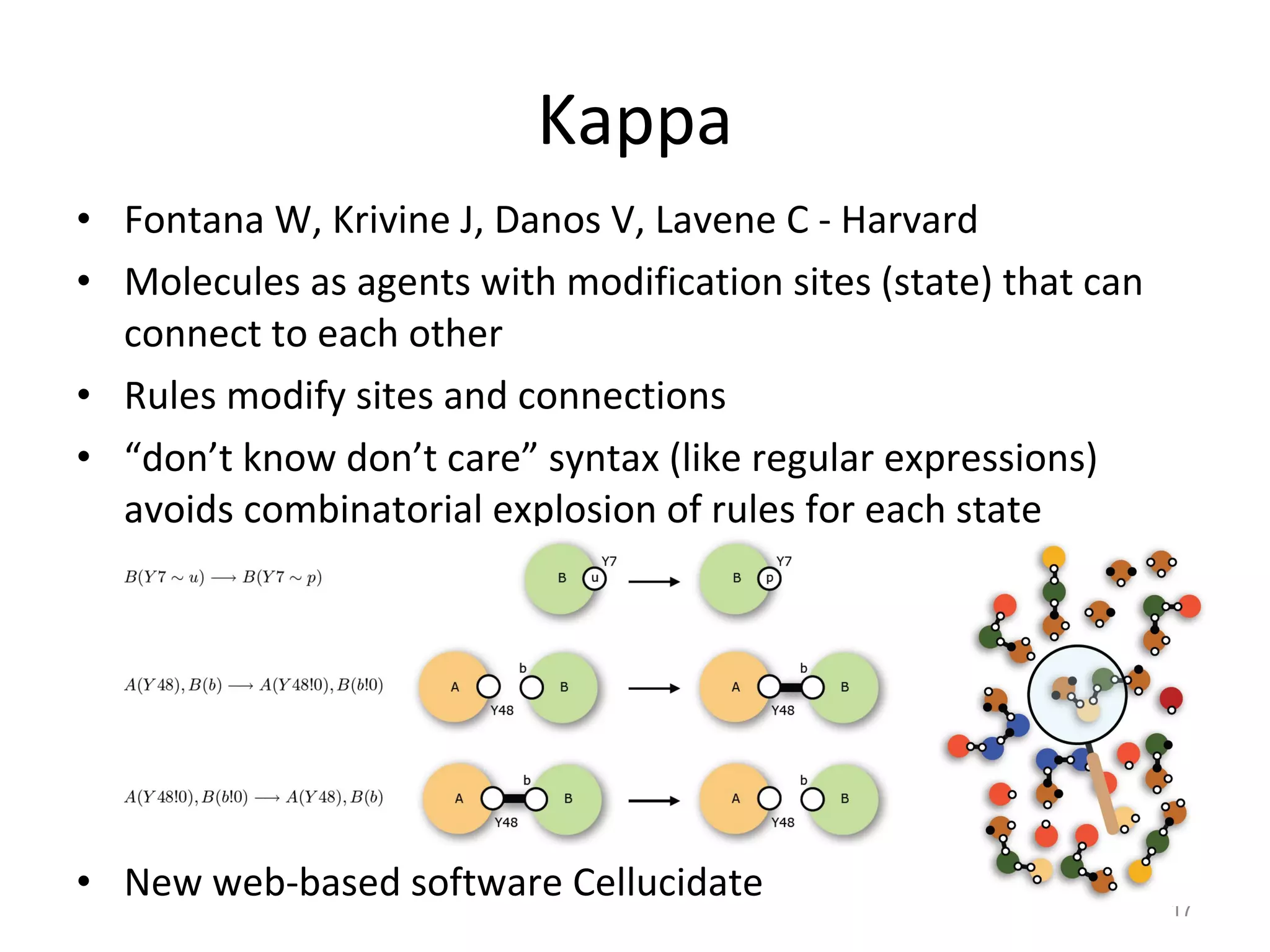
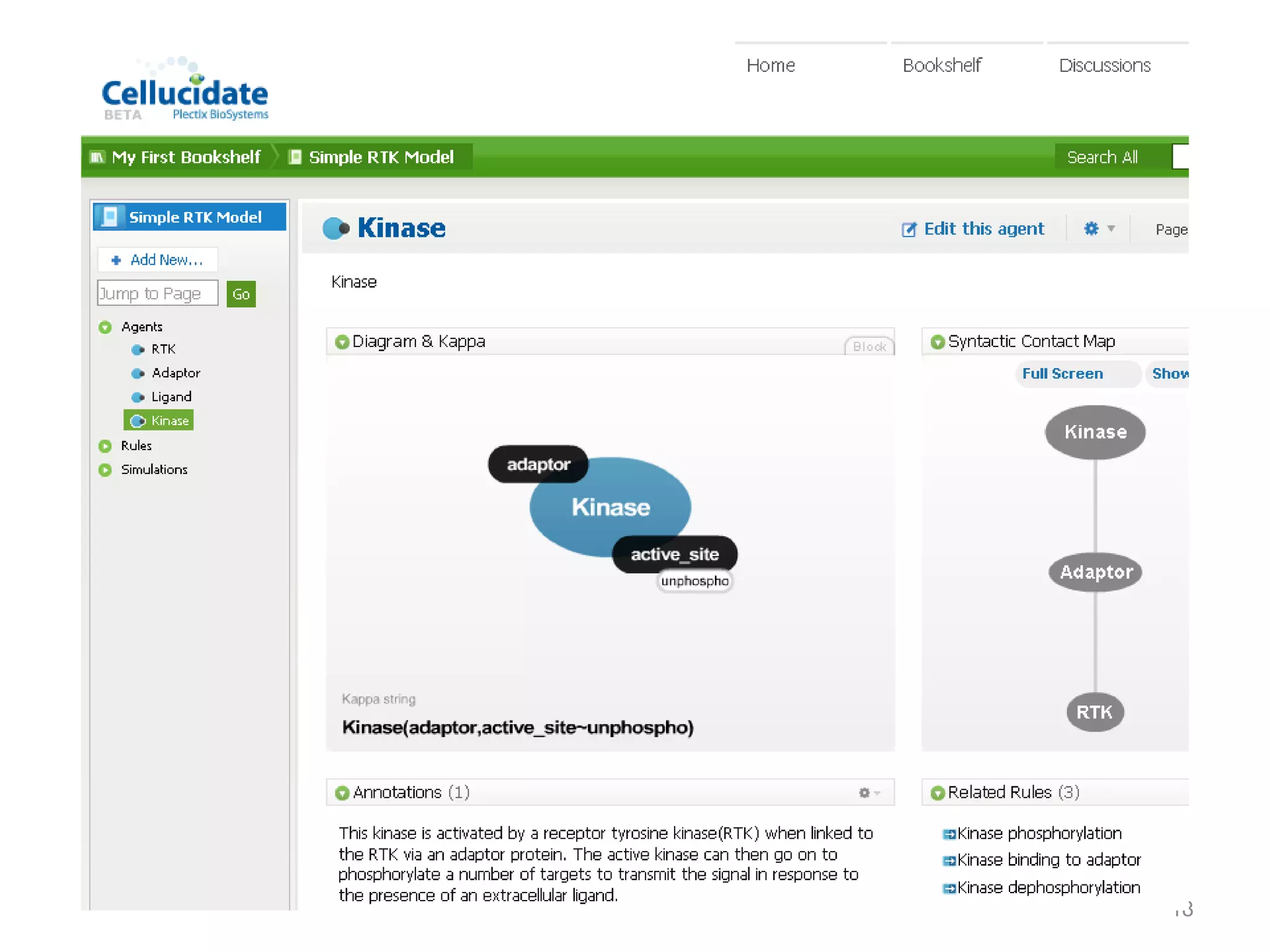
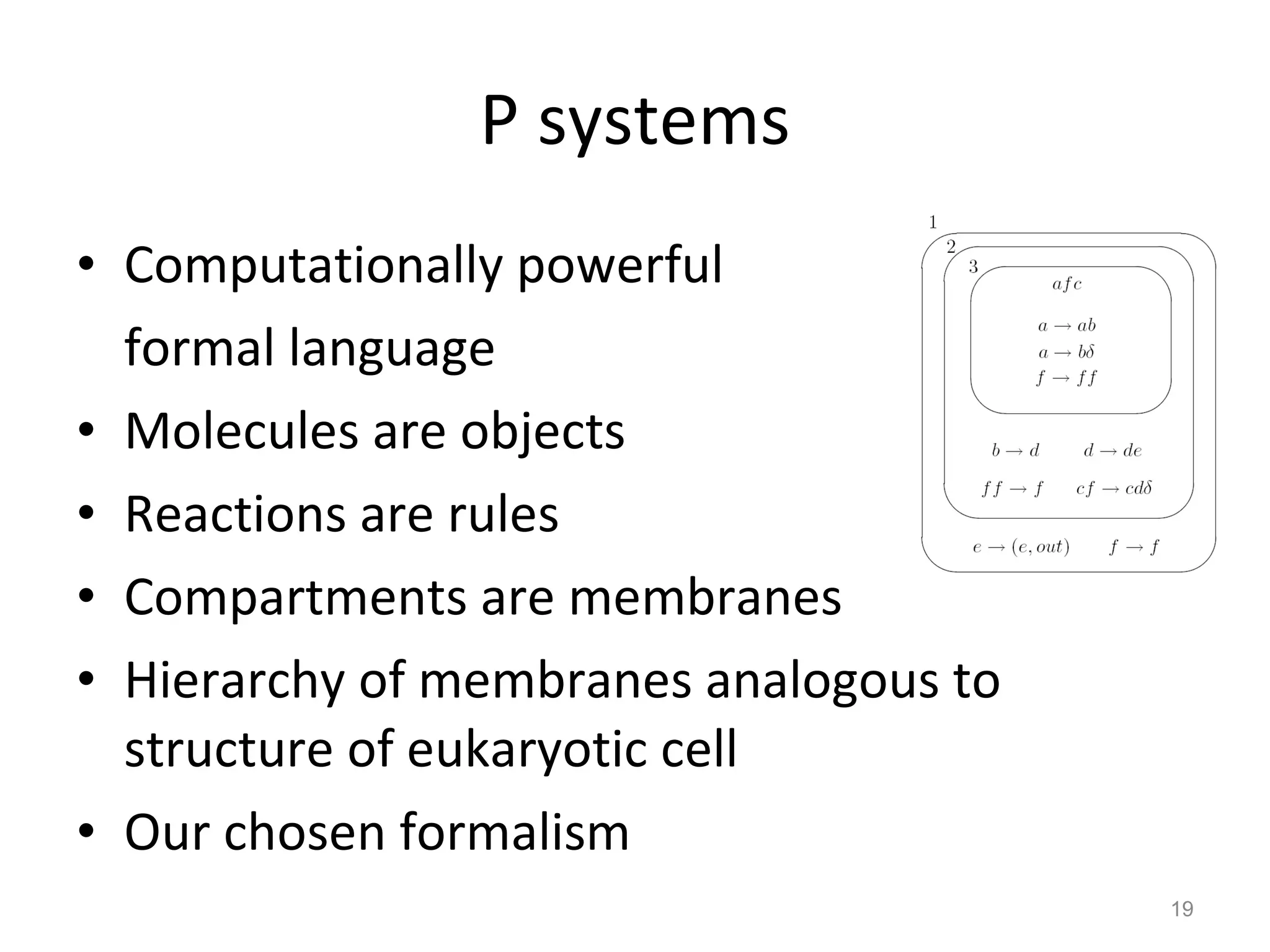
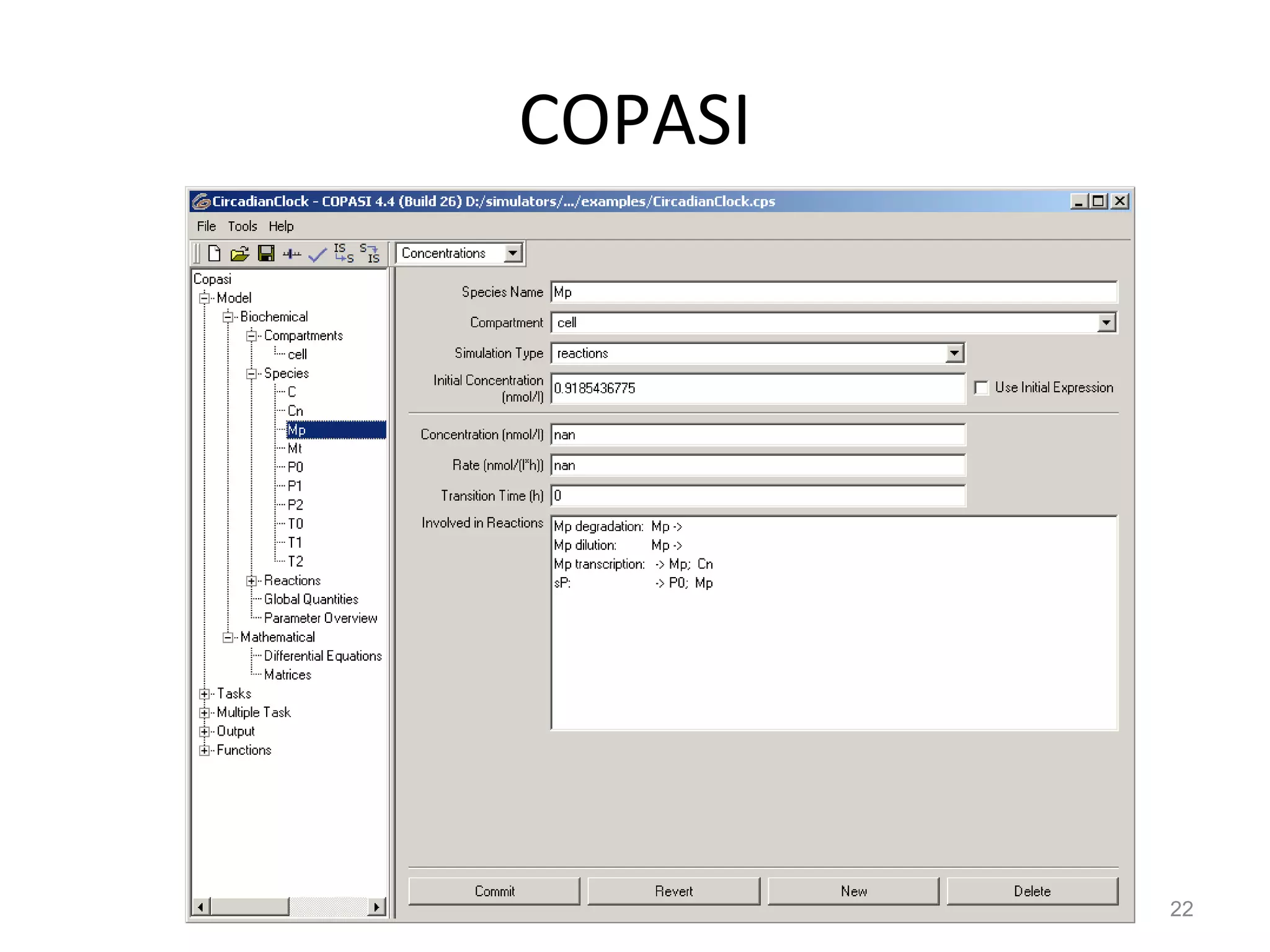
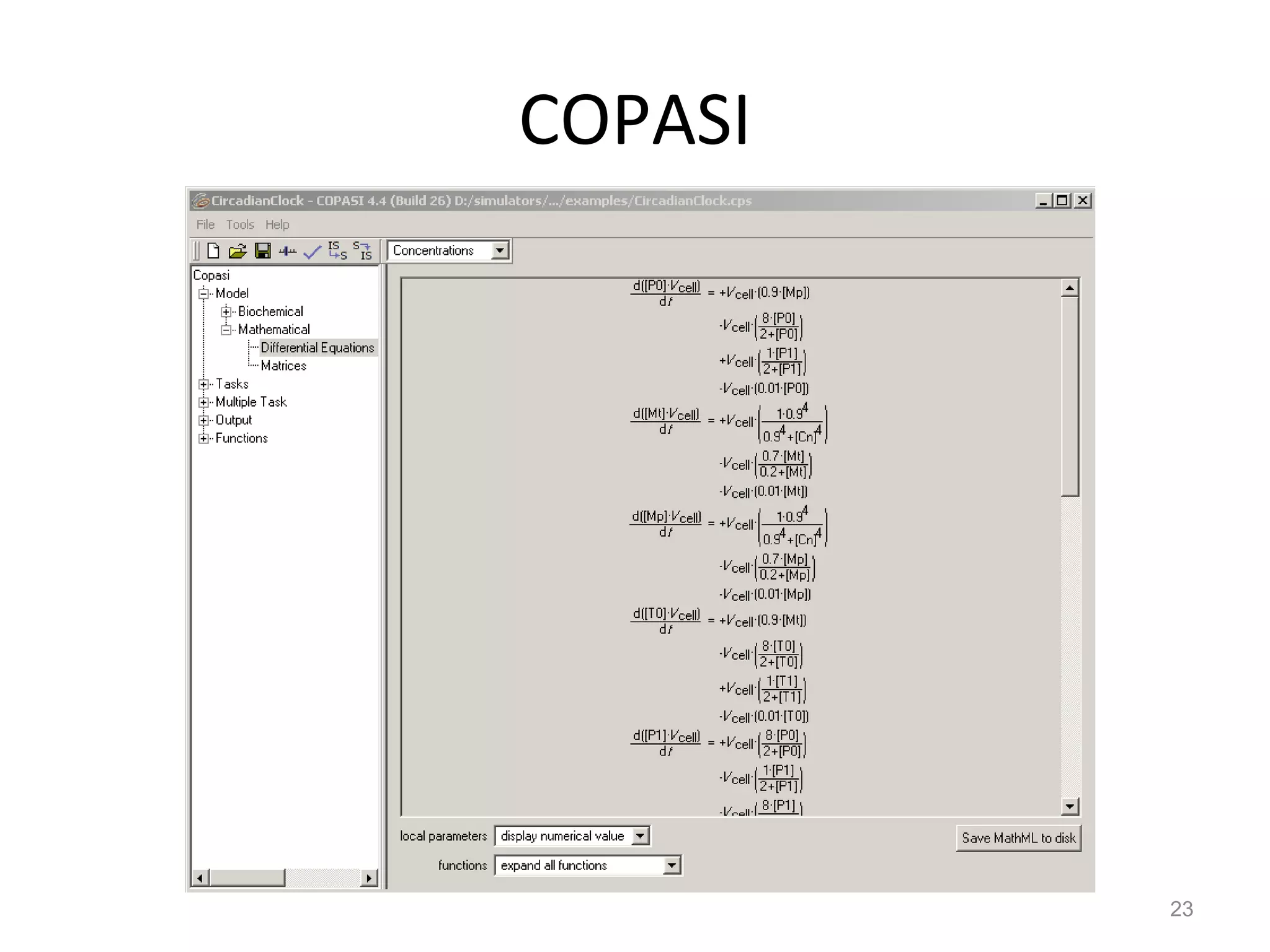
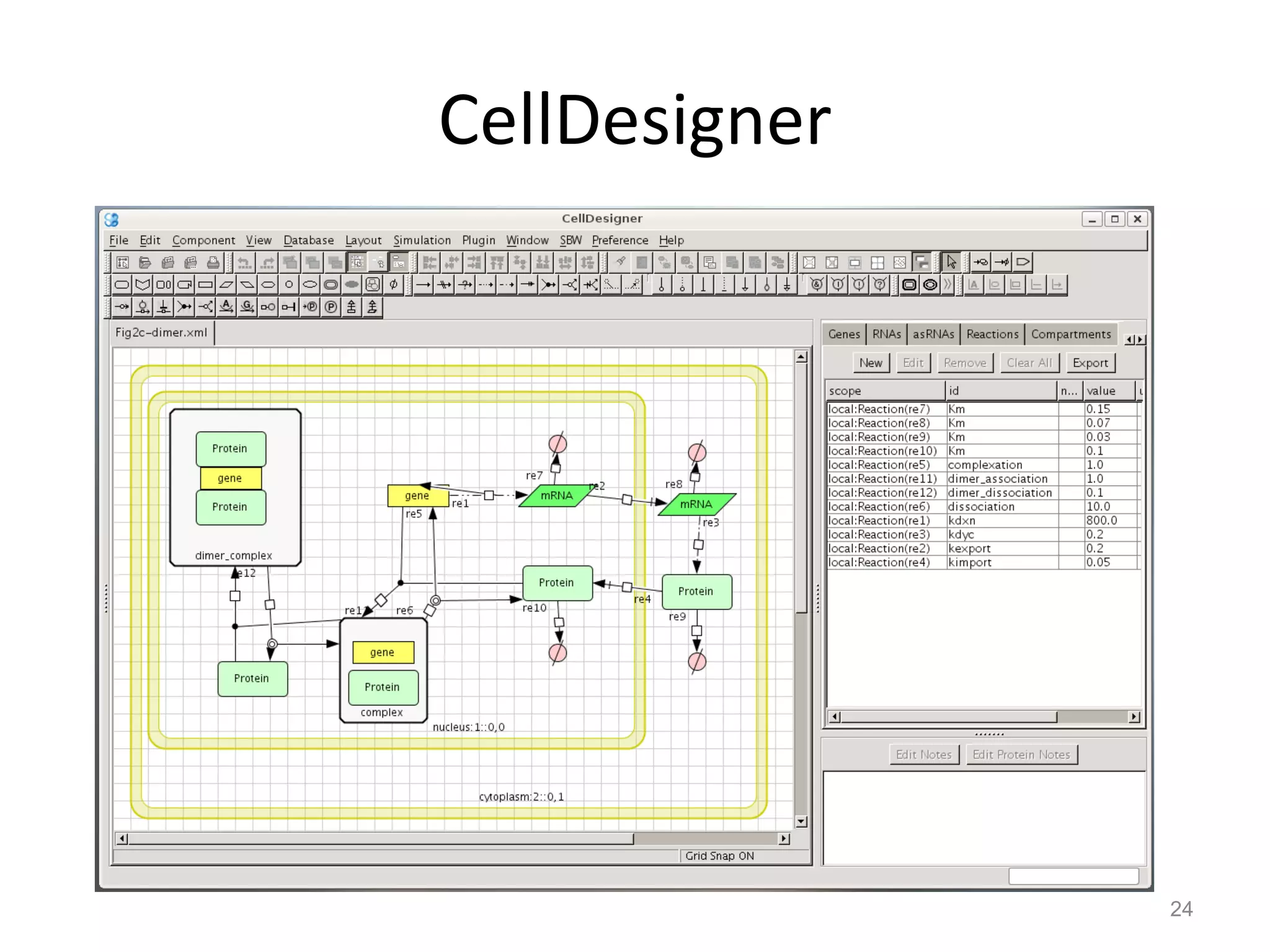
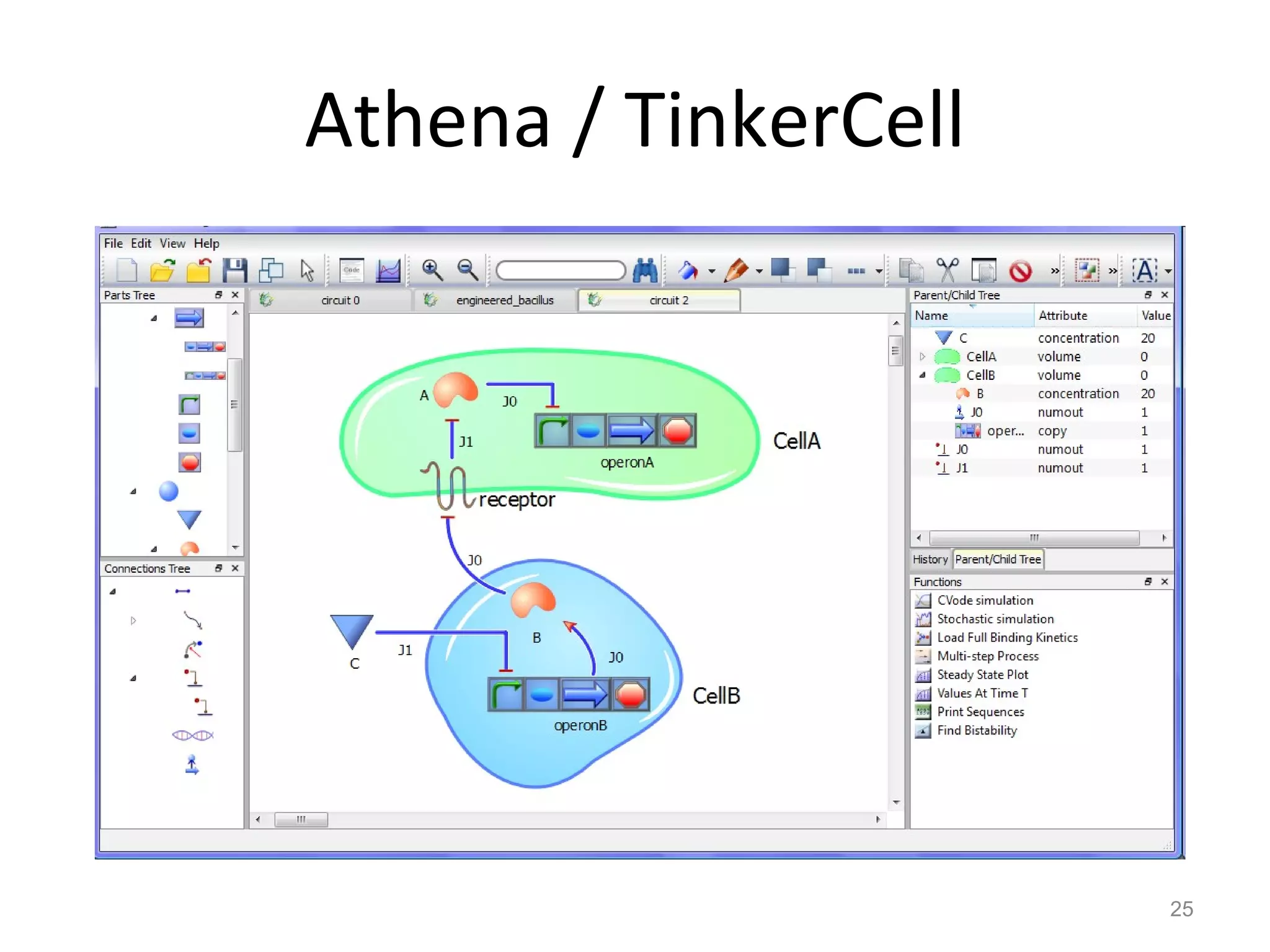
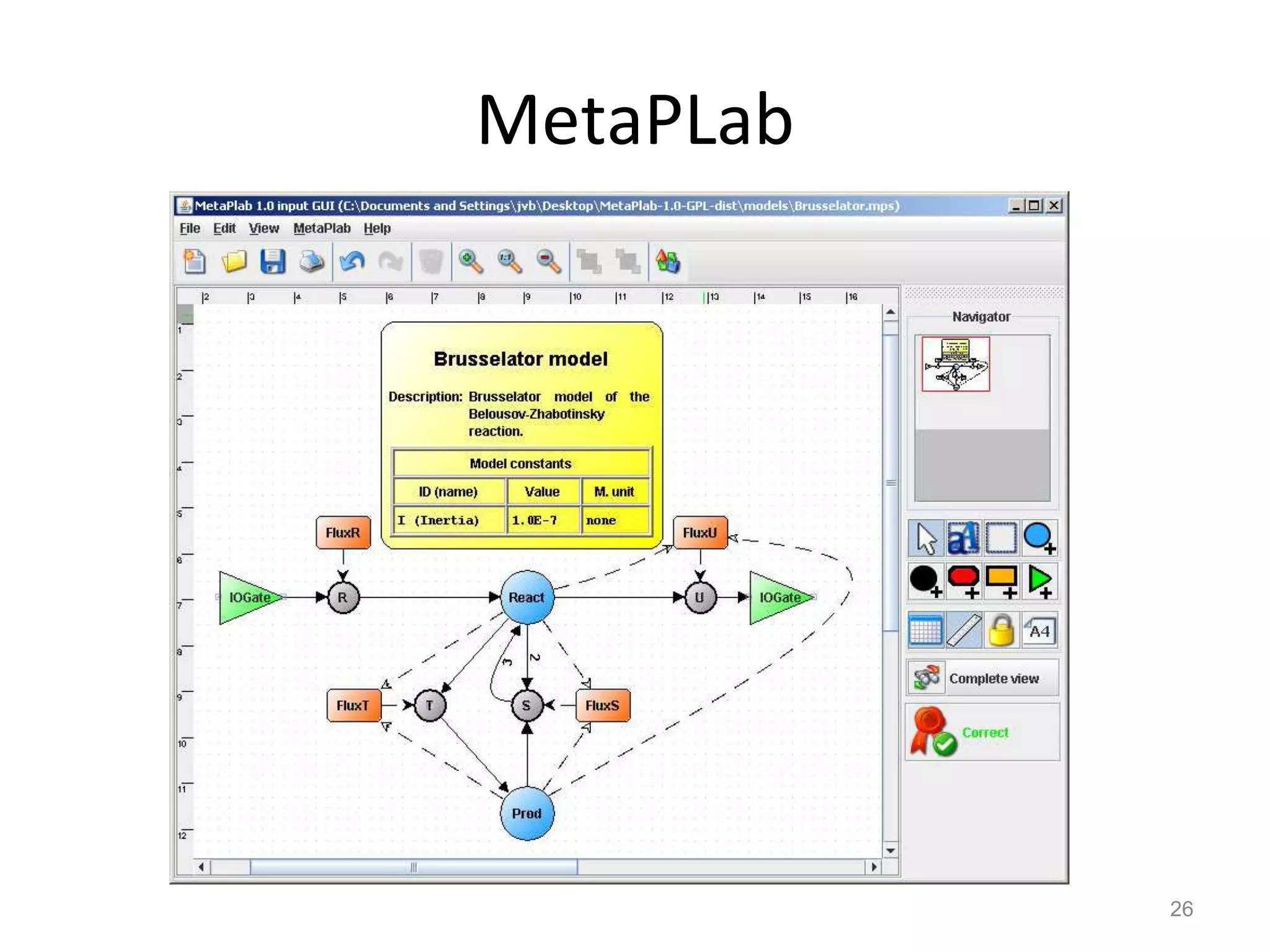
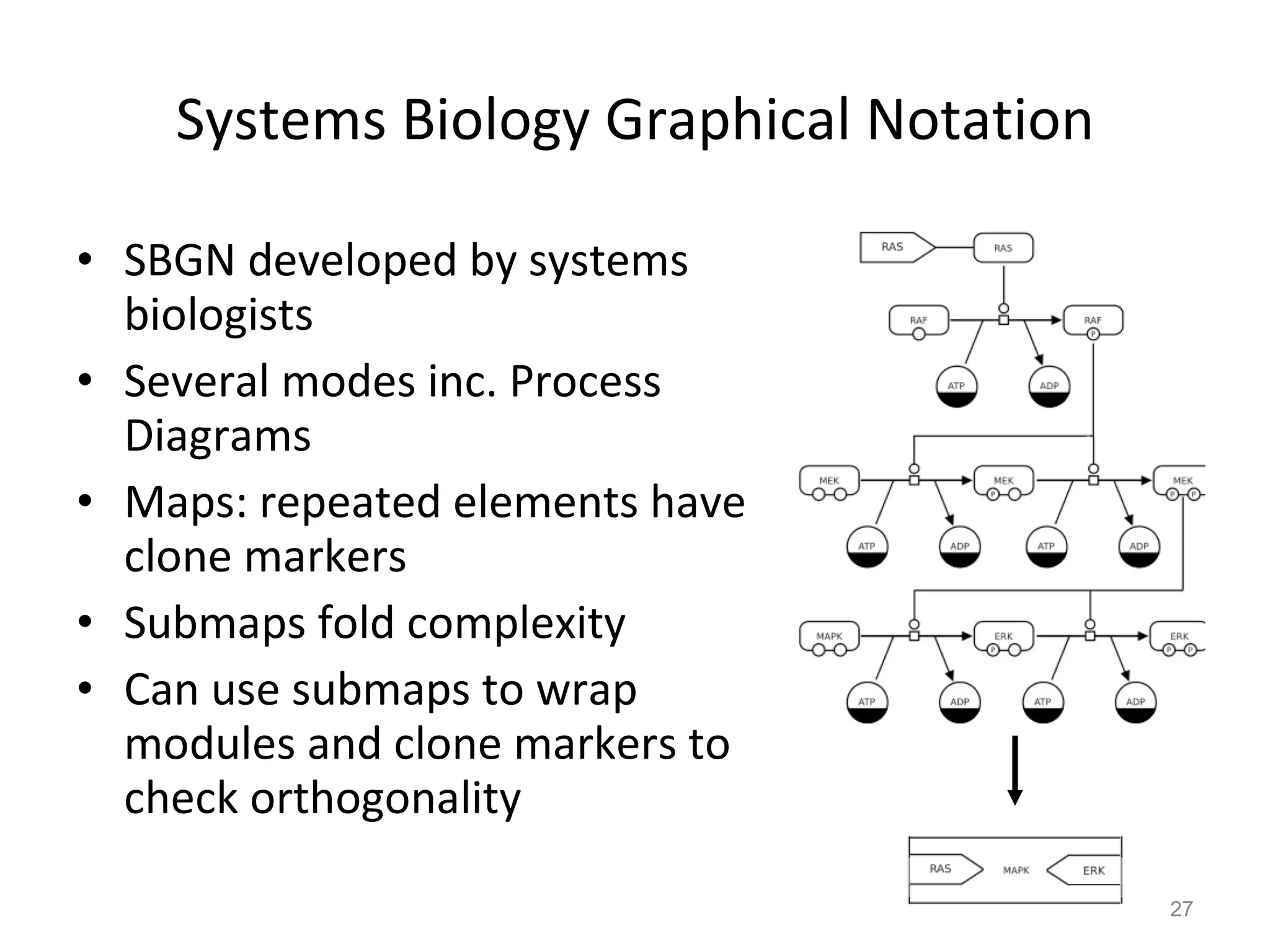
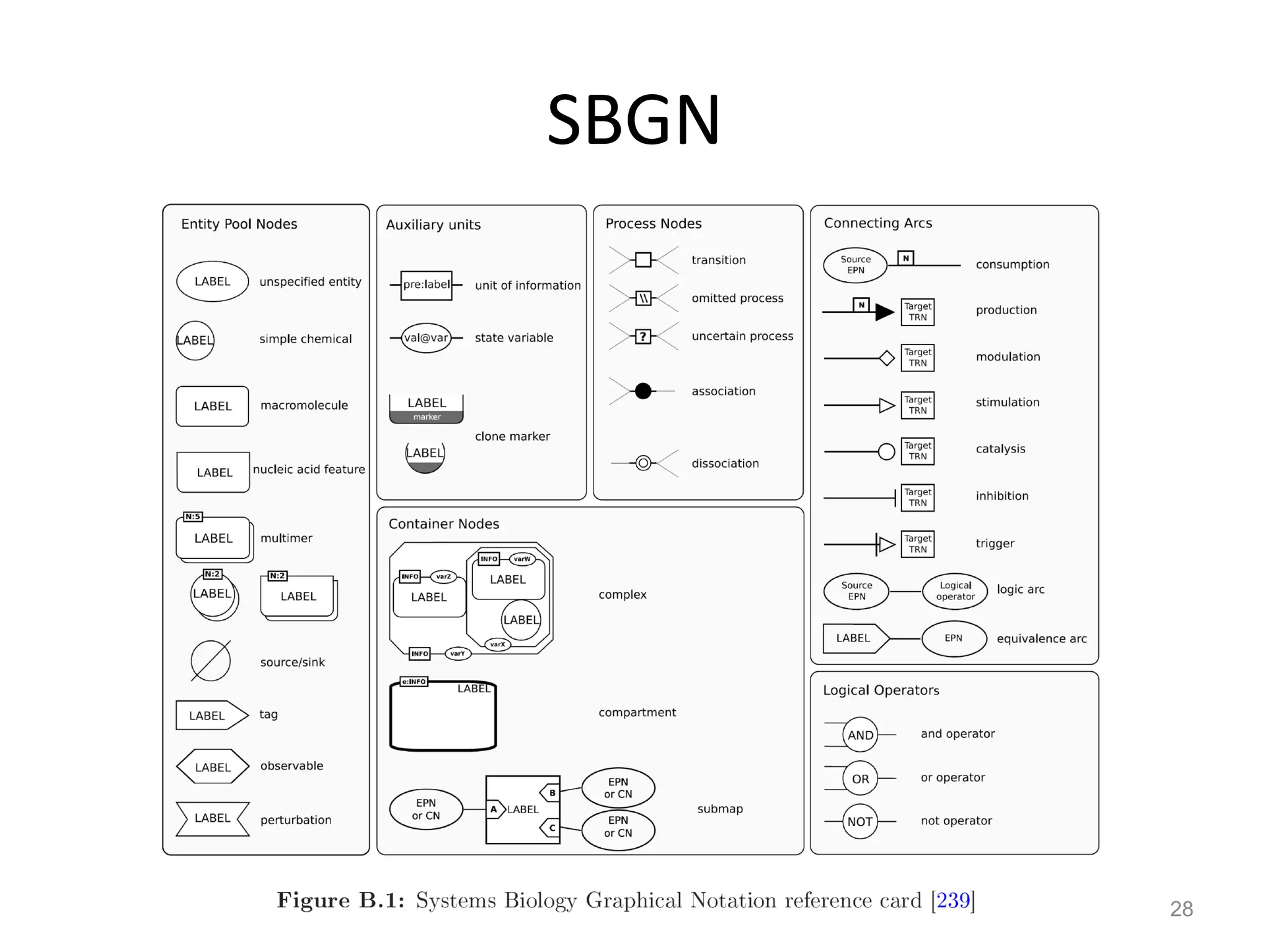
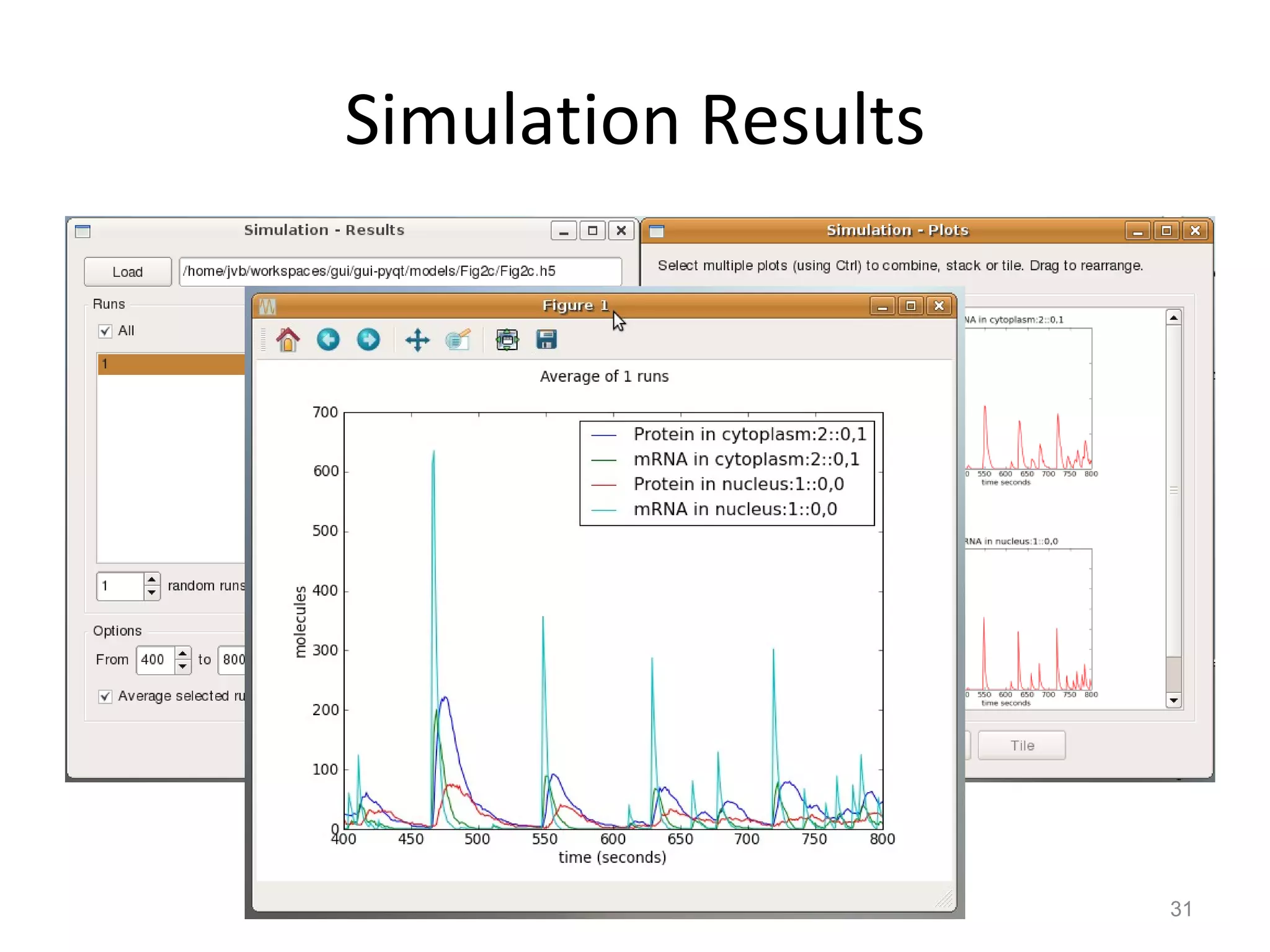
The document discusses the need for a new systems biology modelling environment. It provides context on systems biology and existing modelling approaches and software. It then makes the case that a new modelling environment could improve the user experience for biologists by making models easier to build and refine while allowing for more complex models at larger scales. Key details on existing challenges and the proposed new environment are outlined.