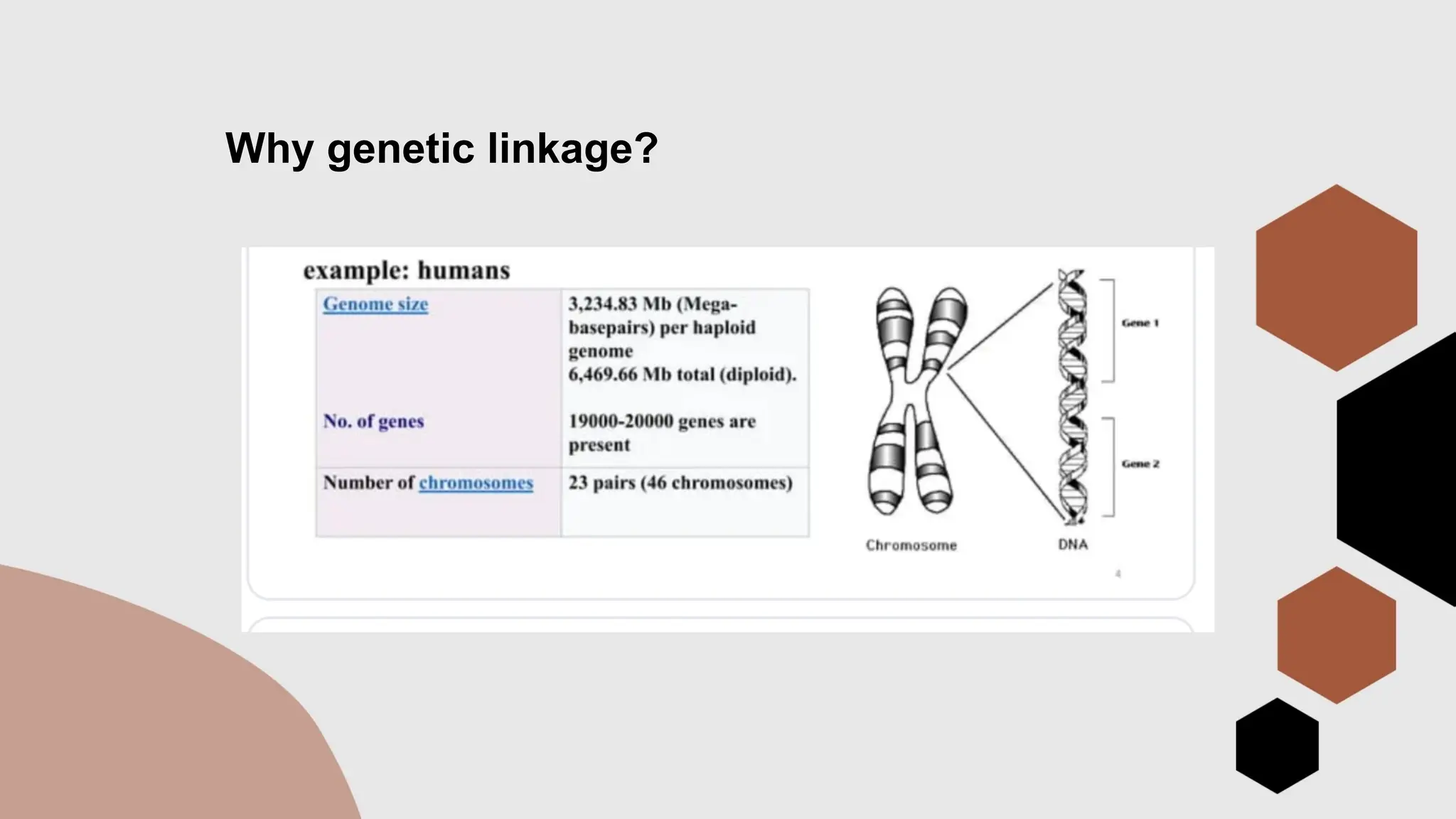
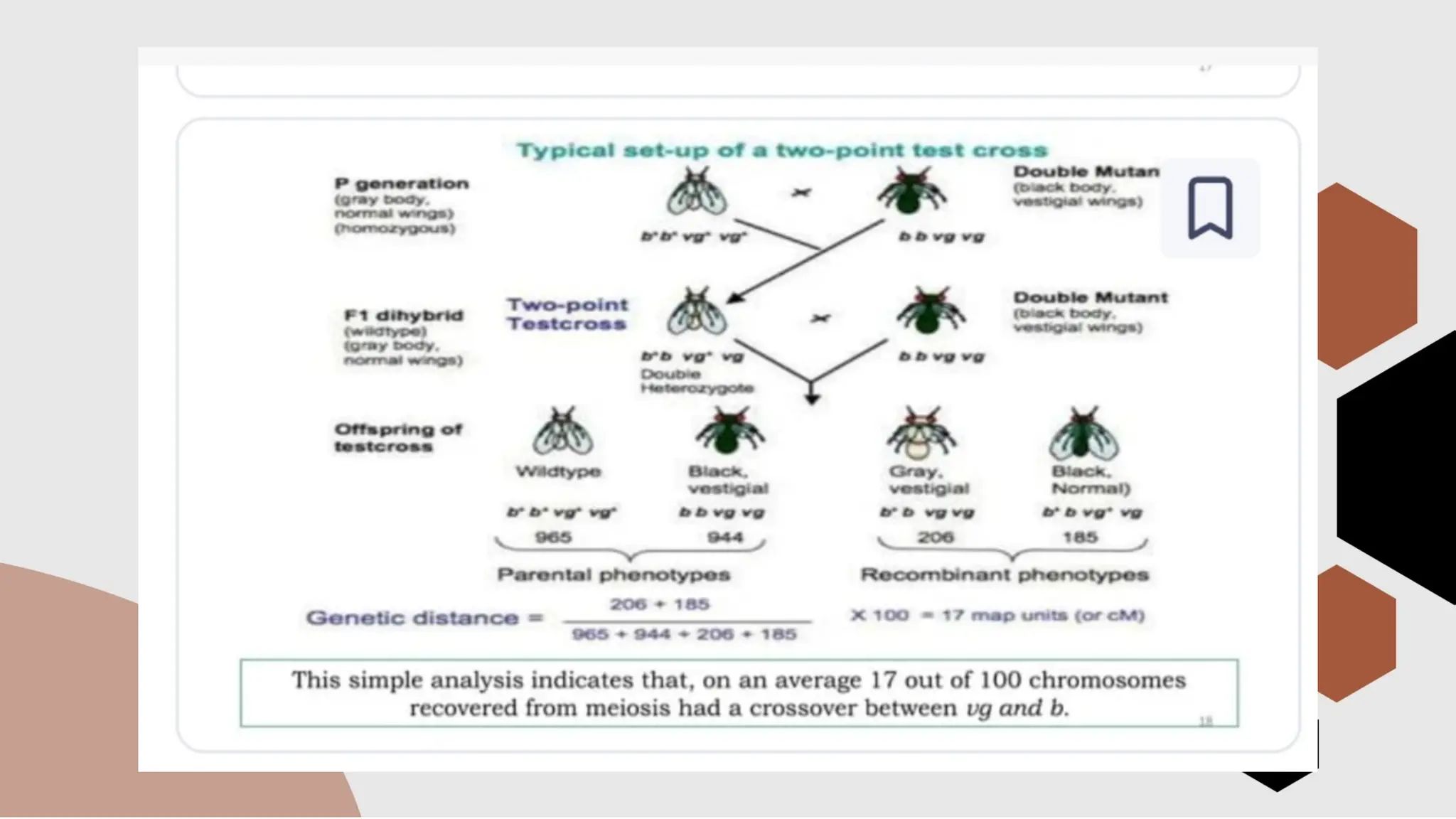
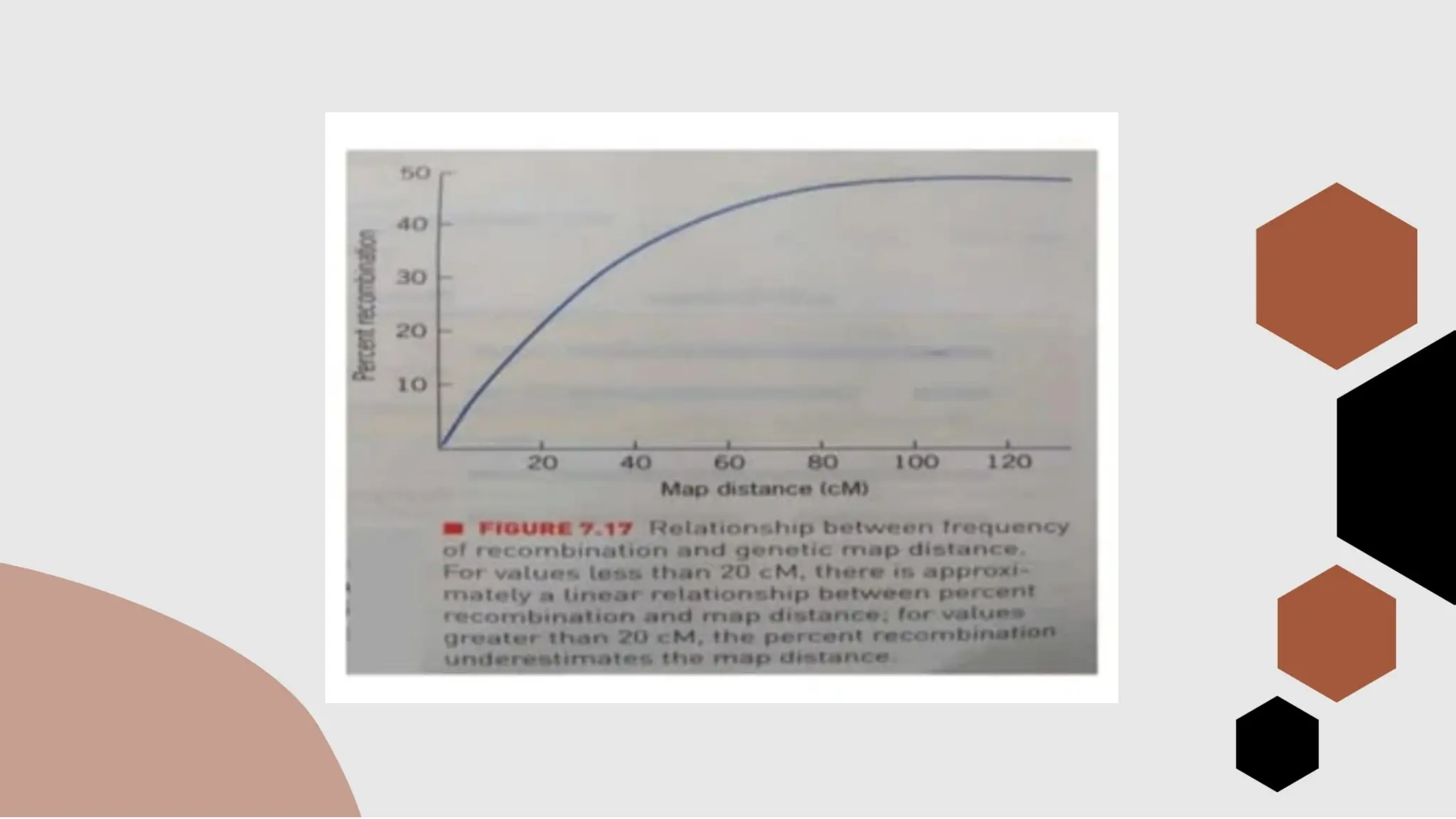
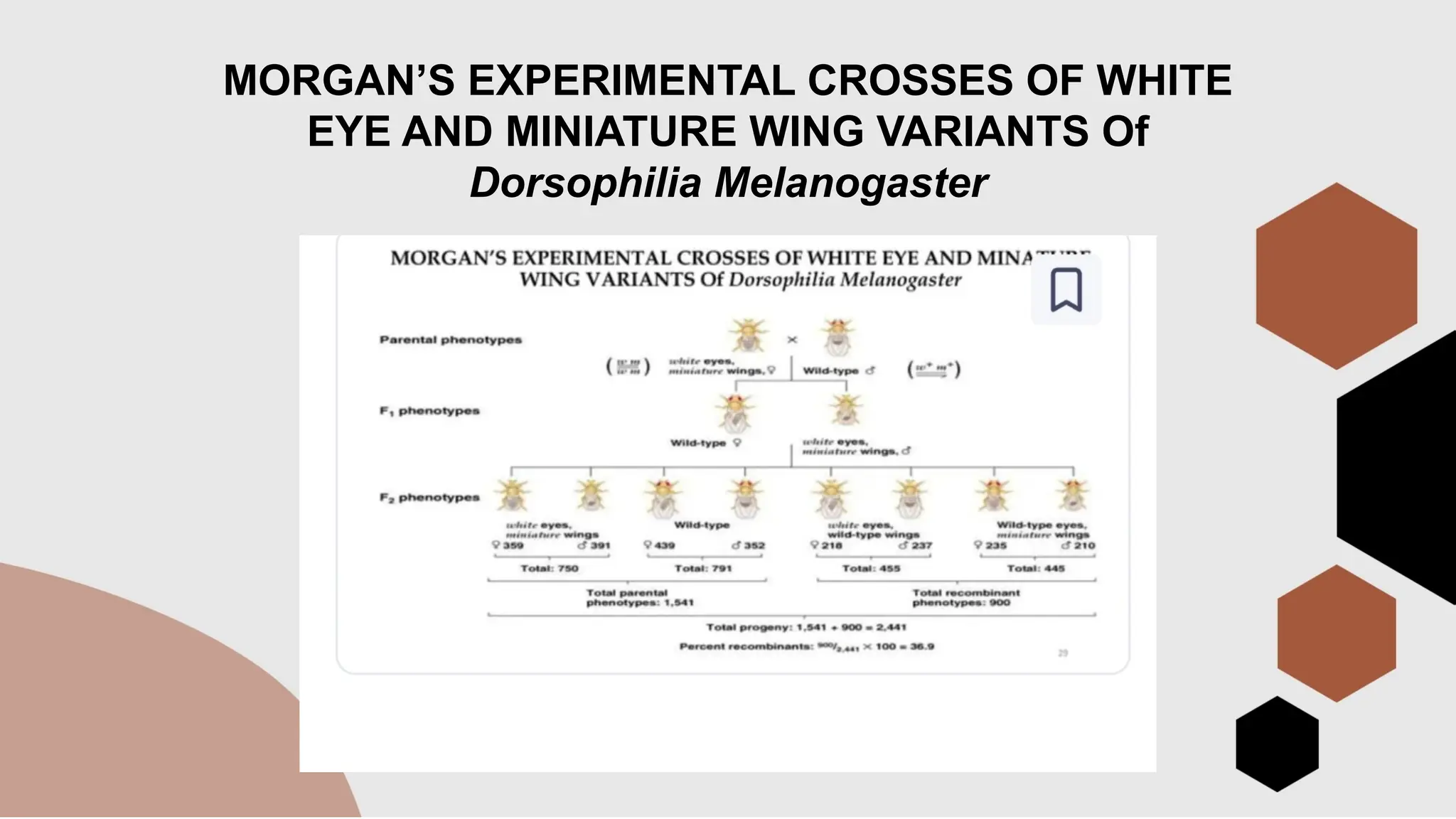
The document discusses genetic linkage, a phenomenon where DNA sequences close together on a chromosome tend to be inherited together, first discovered by Bateson and Punnett in 1906. It explains the concepts of recombination, factors affecting recombination frequency, types of linkage, and methods of chromosome mapping, highlighting experiments by Creighton, McClintock, and Morgan. The document concludes with an analysis of linkage groups and genetic mapping techniques, emphasizing the relationship between recombination and the inheritance of traits in organisms.