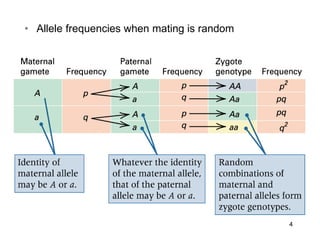
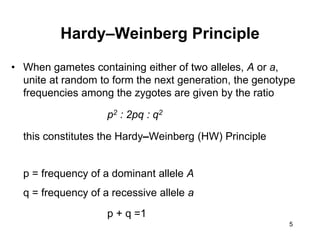
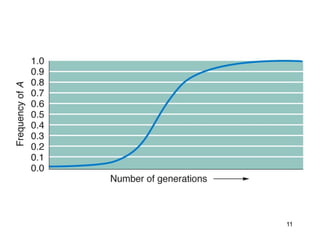
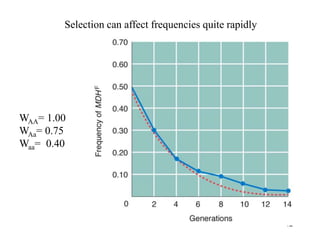
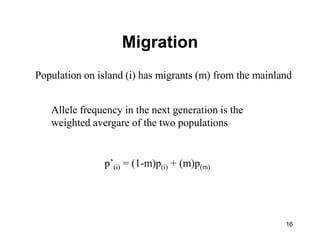
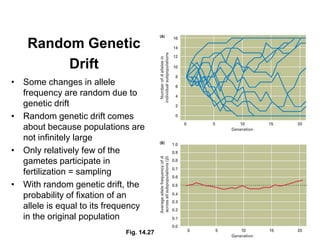
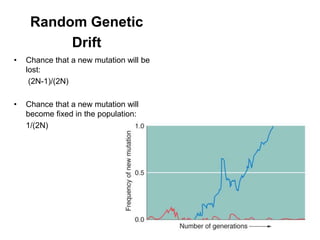
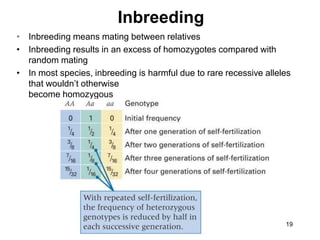
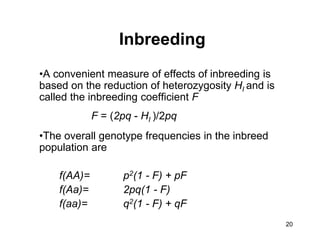
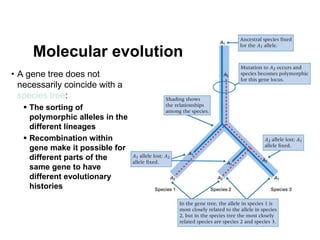
The document discusses population genetics, defining key concepts such as populations, gene pools, allele frequencies, and the Hardy-Weinberg principle, which states genetic variation remains constant under specific conditions. It covers the effects of mutation, natural selection, migration, and random genetic drift on allele frequencies, as well as the implications of inbreeding and heterozygote superiority. Additionally, it addresses molecular evolution, emphasizing the comparison of genetic material across species and the role of gene trees in understanding evolutionary relationships.