The Infobiotics BioProgramming Language & Workbench provides a computer-aided design environment for synthetic biology. It integrates simulation, verification, and compilation capabilities through an iterative workflow. The Infobiotics Language (IBL) allows users to define synthetic biology parts, rules, and devices. IBL supports abstraction, encapsulation, and hierarchical organization. The workbench performs stochastic simulation, model checking for verification, and biomatter compilation to generate DNA sequences. It aims to enable more reliable engineering of synthetic biological circuits.








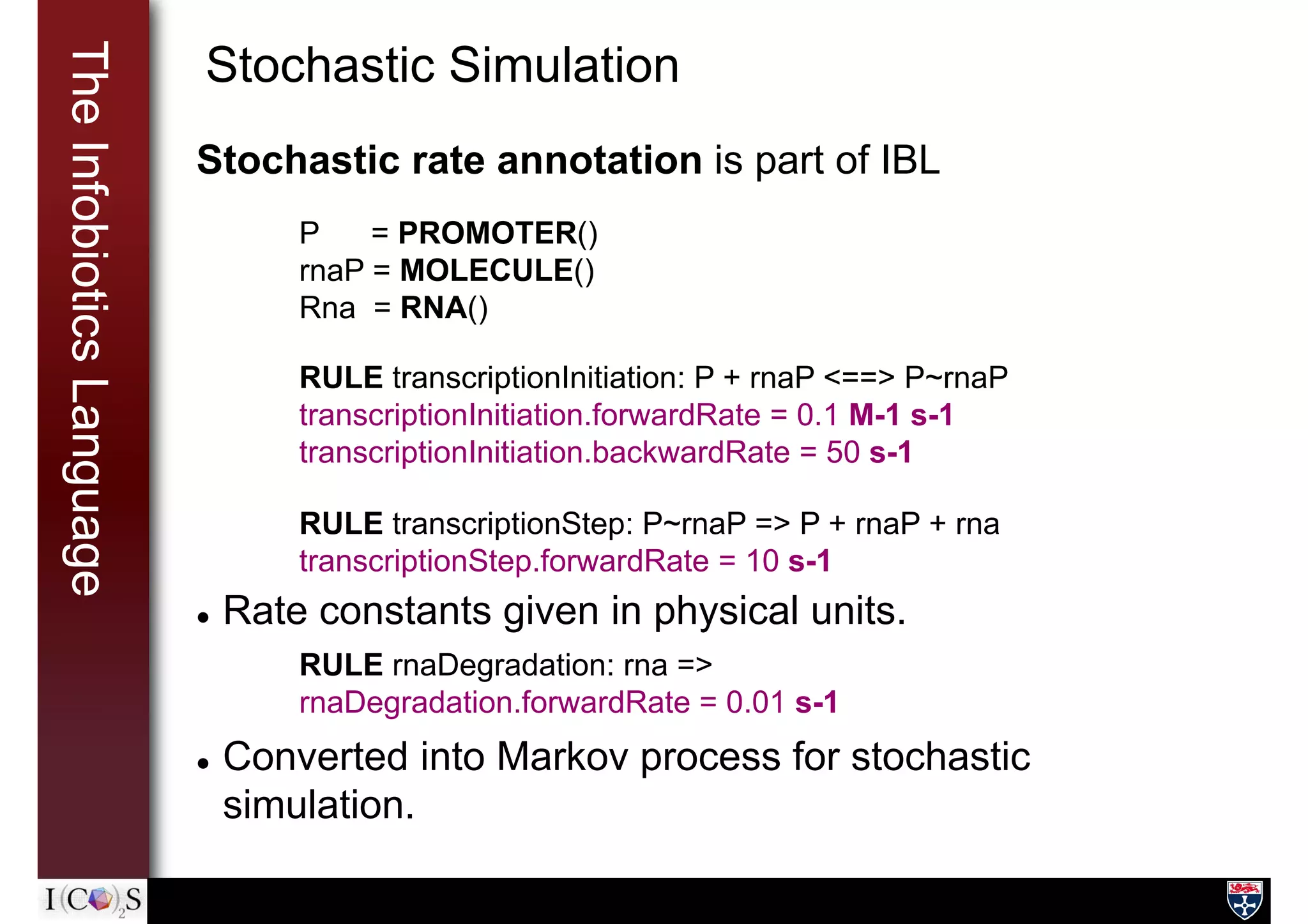
![TheInfobioticsLanguage Putting it all together: Devices
A device in the SB sense: a continuous piece of DNA
DEVICE(
parts = [lacI, pLs1con, pTrc2, cI, gfpmut3],
input = [aTc, IPTG], output = [LacI, CI, GFPmut3]
) {
mrna_LacI = RNA() // a local variable
// PROCESSES and RULES here
}
GFPmut3cI
LacI pTrc2
pLs1con
IPTG
aTc
collection of parts,
rules, and processes
that characterize the
piece of DNA](https://image.slidesharecdn.com/l2-theibw-170214161143/75/The-Infobiotics-workbench-10-2048.jpg)




![TheInfobioticsLanguage Verification through Model Checking
Verification statements are part of IBL
l Natural language like syntax
l Properties obtained from mining biological literature
l Can be converted into temporal logic clauses and fed
into different model checkers (PRISM, NuSVM, MS2)
VERIFY [ GFP > 0 uM ] EVENTUALLY HOLDS
VERIFY [ GFP > 0 uM ] EVENTUALLY HOLDS WITH PROBABILITY > 0.9
VERIFY [ GFP > 0 uM ] ALWAYS HOLDS
VERIFY [ GFP > 2*RFP ] NEVER HOLDS
VERIFY [ GFP > 0 uM ] HOLDS WITHIN 60 s
VERIFY [ AHL > 0 uM ] IS FOLLOWED BY [ GFP > 0 uM ]
VERIFY [ GFP > 0 uM ] EVENTUALLY HOLDS,
UNTIL THEN [ NahR = 0 uM ] HOLDS](https://image.slidesharecdn.com/l2-theibw-170214161143/75/The-Infobiotics-workbench-16-2048.jpg)
![TheInfobioticsWorkbench Verification through Model Checking
Verification is performed in the defining context:
Allows scaling of model checking to systems with many
components.
Verification of RULEs
Verification of CELLs
Verification of DEVICEs
define myDevice typeof DEVICE() {
// RULEs tested during verification
VERIFY [ GFP > 0 uM ] EVENTUALLY HOLDS
}
// other DEVICEs and RULEs, not tested during verification](https://image.slidesharecdn.com/l2-theibw-170214161143/75/The-Infobiotics-workbench-17-2048.jpg)

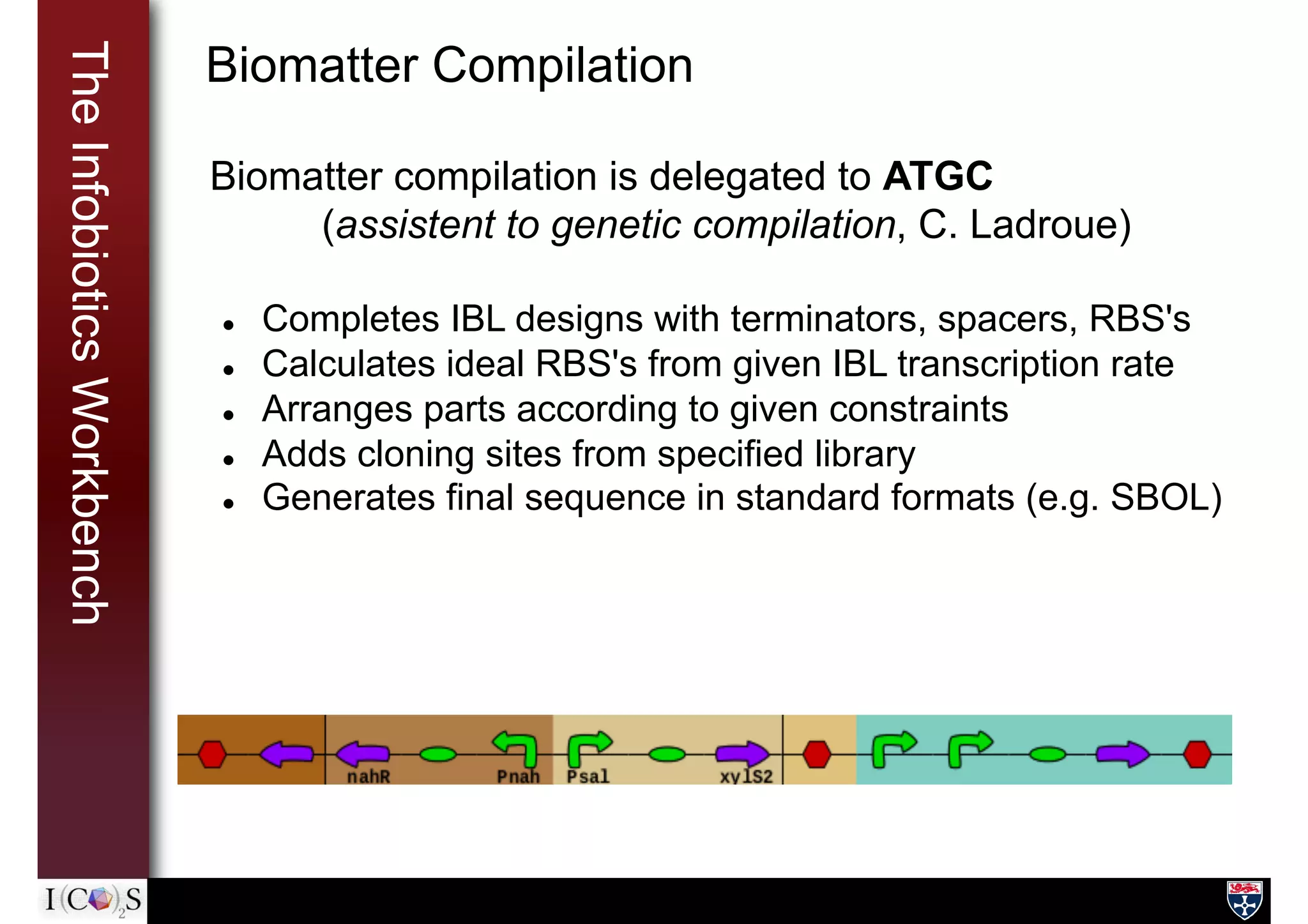
![TheInfobioticsLanguage Biomatter Compilation
Biomatter compilation directives are part of IBL
Sequence information can be pulled in from standard
repositories
(e.g. biobricks, virtualParts), in-house databases, or defined
manually.
PnahR = PROMOTER(URI = "biobricks://BB_0132")
nahR = GENE(URI = "biobricks://BB_08431")
lasR = GENE(sequence= "TACGTTGACCA...")
myDevice = DEVICE(parts=[PnahR, nahR, lasR], output=[NahR, LasR]) {
ATCG ARRANGE nahR lasR
// PROCESSES and RULES here
}
ATCG DEVICE myDevice CLONING SITES: 2](https://image.slidesharecdn.com/l2-theibw-170214161143/75/The-Infobiotics-workbench-19-2048.jpg)