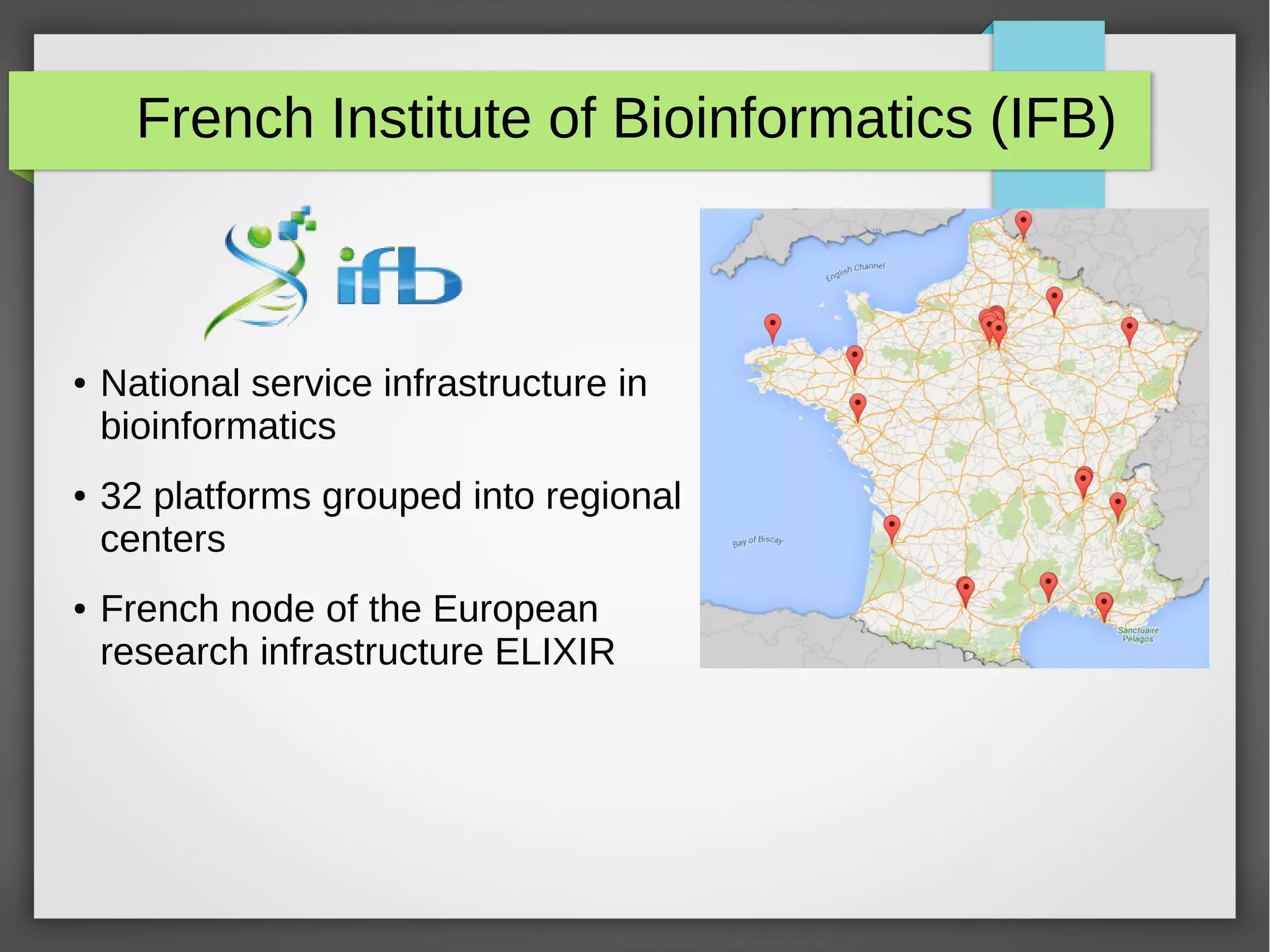
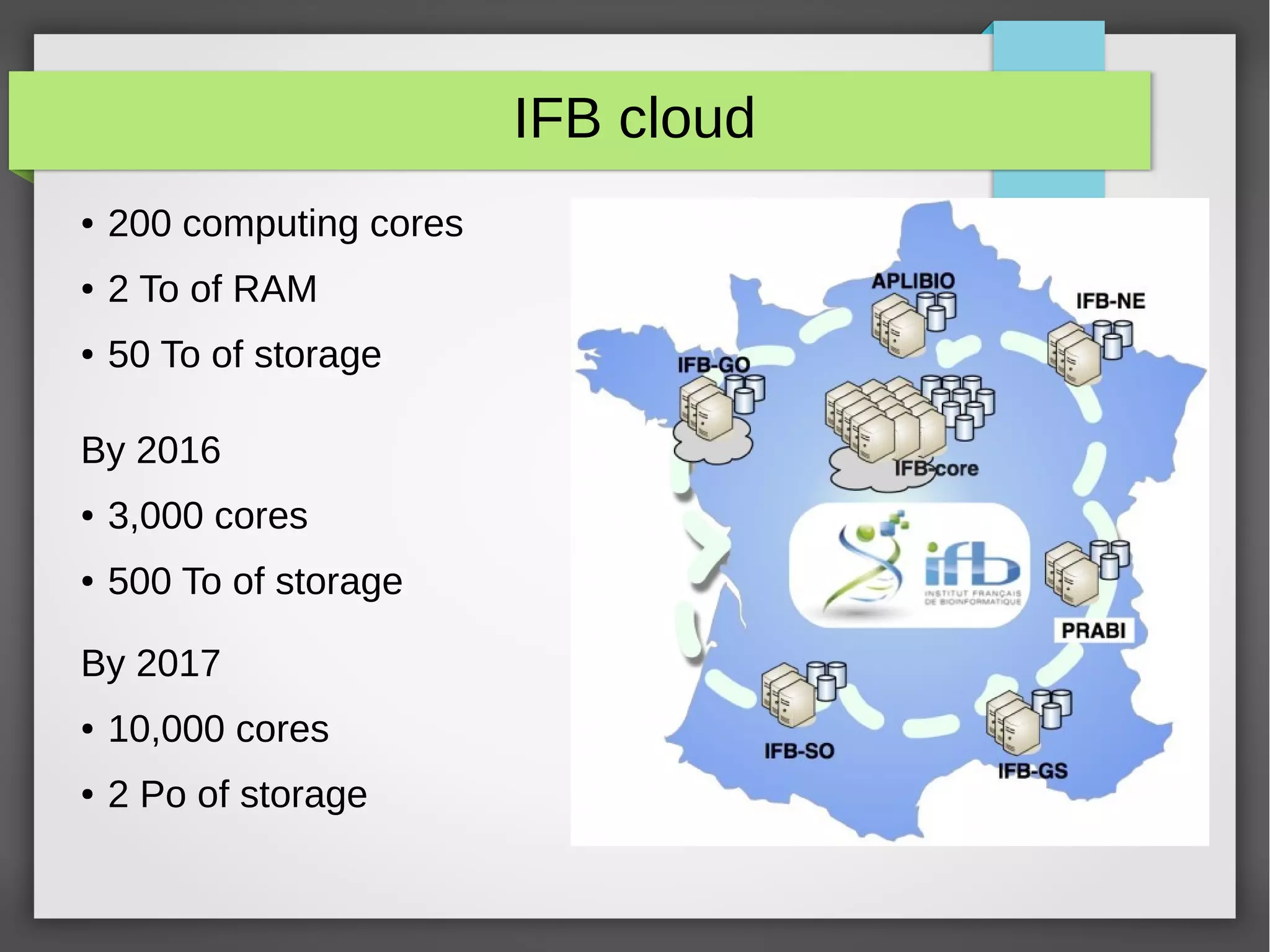
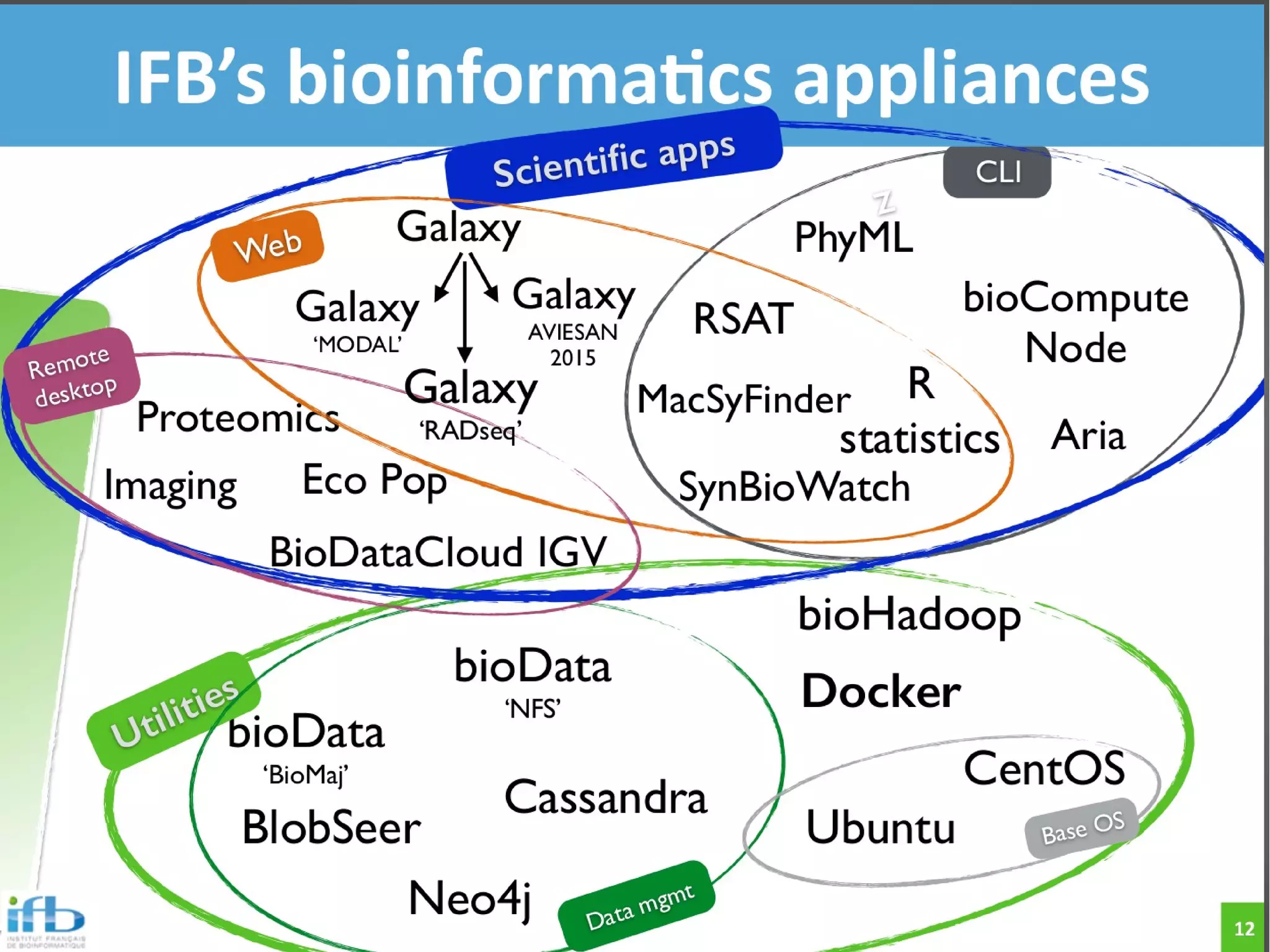
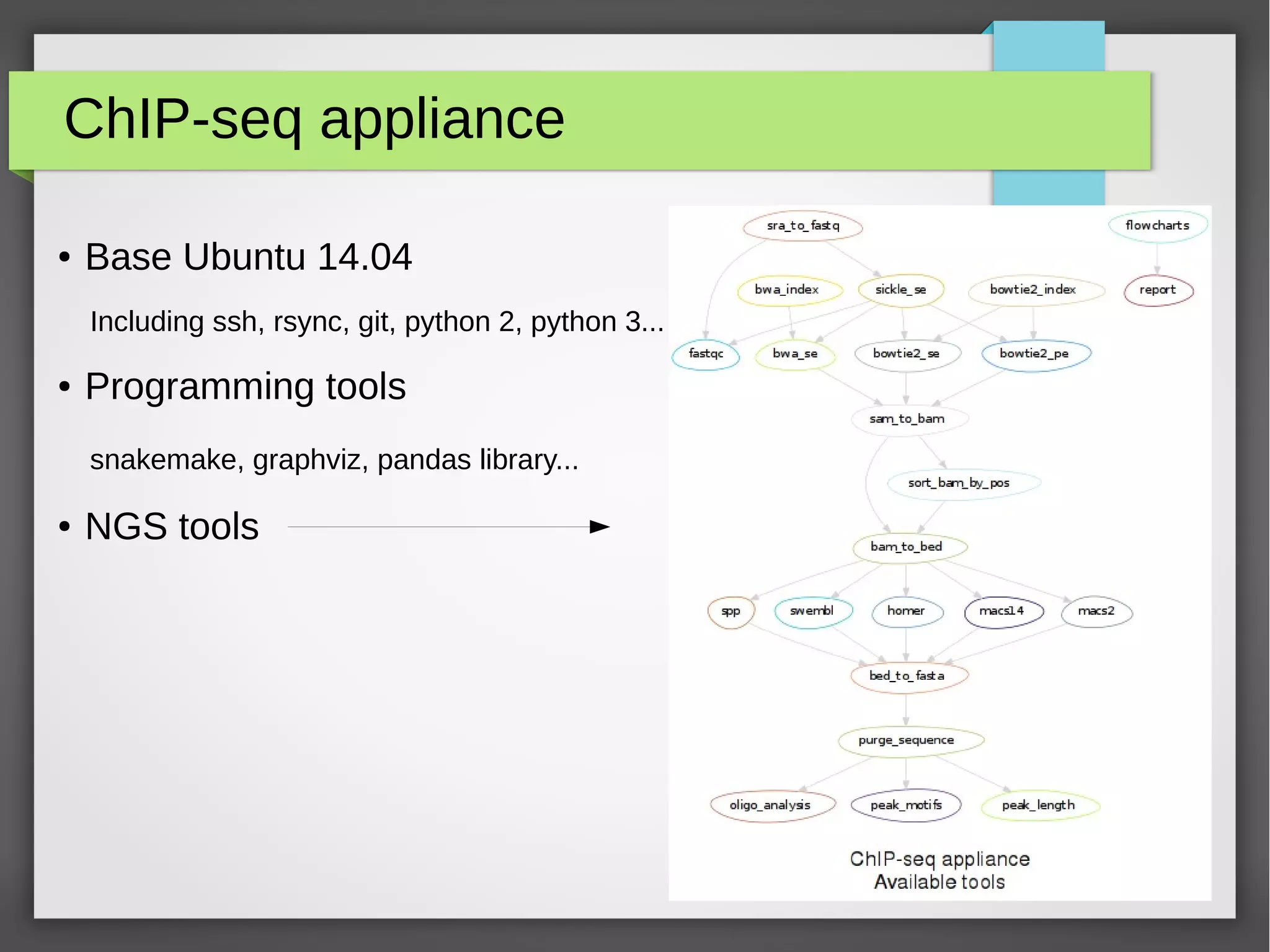
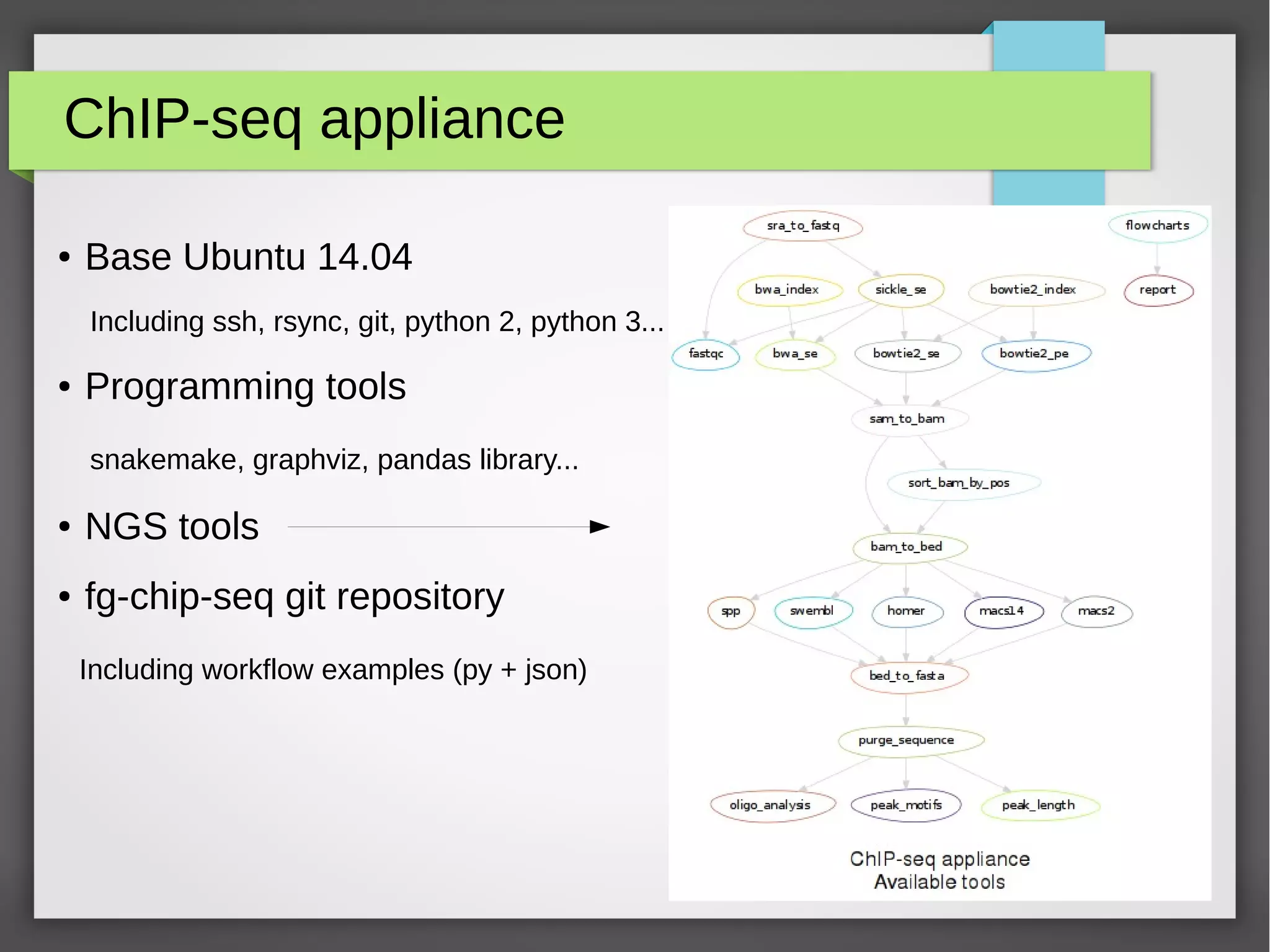
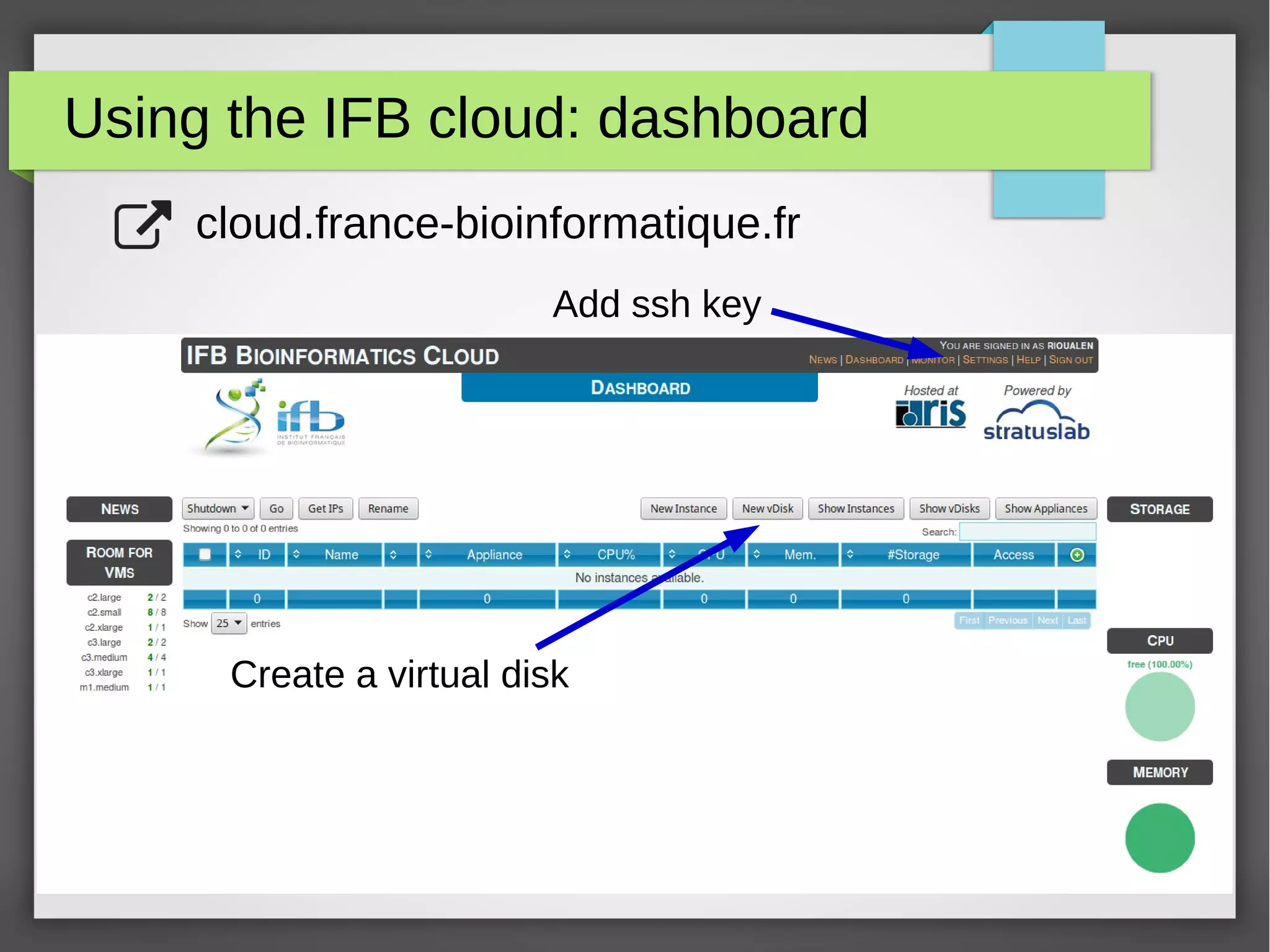
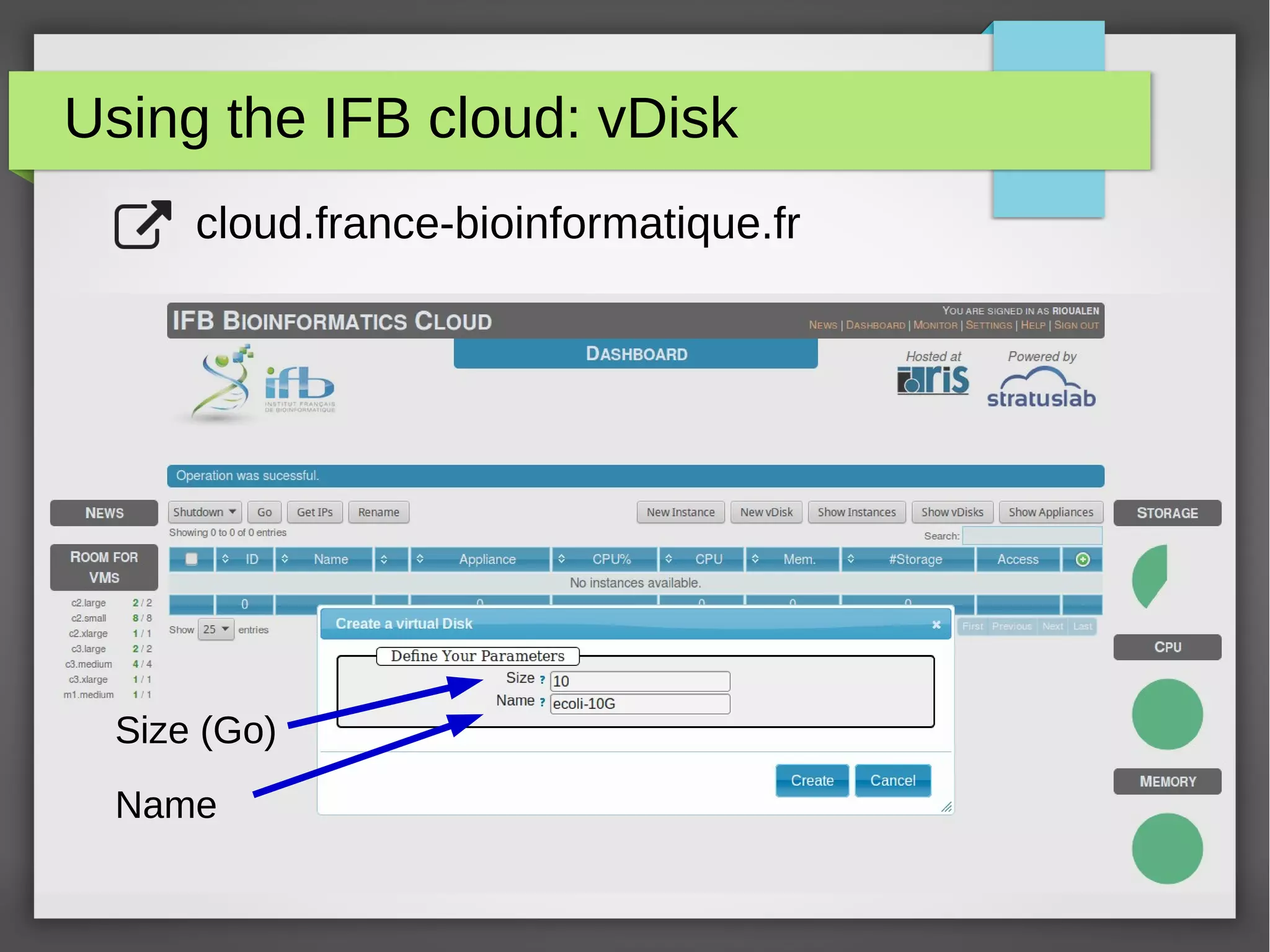
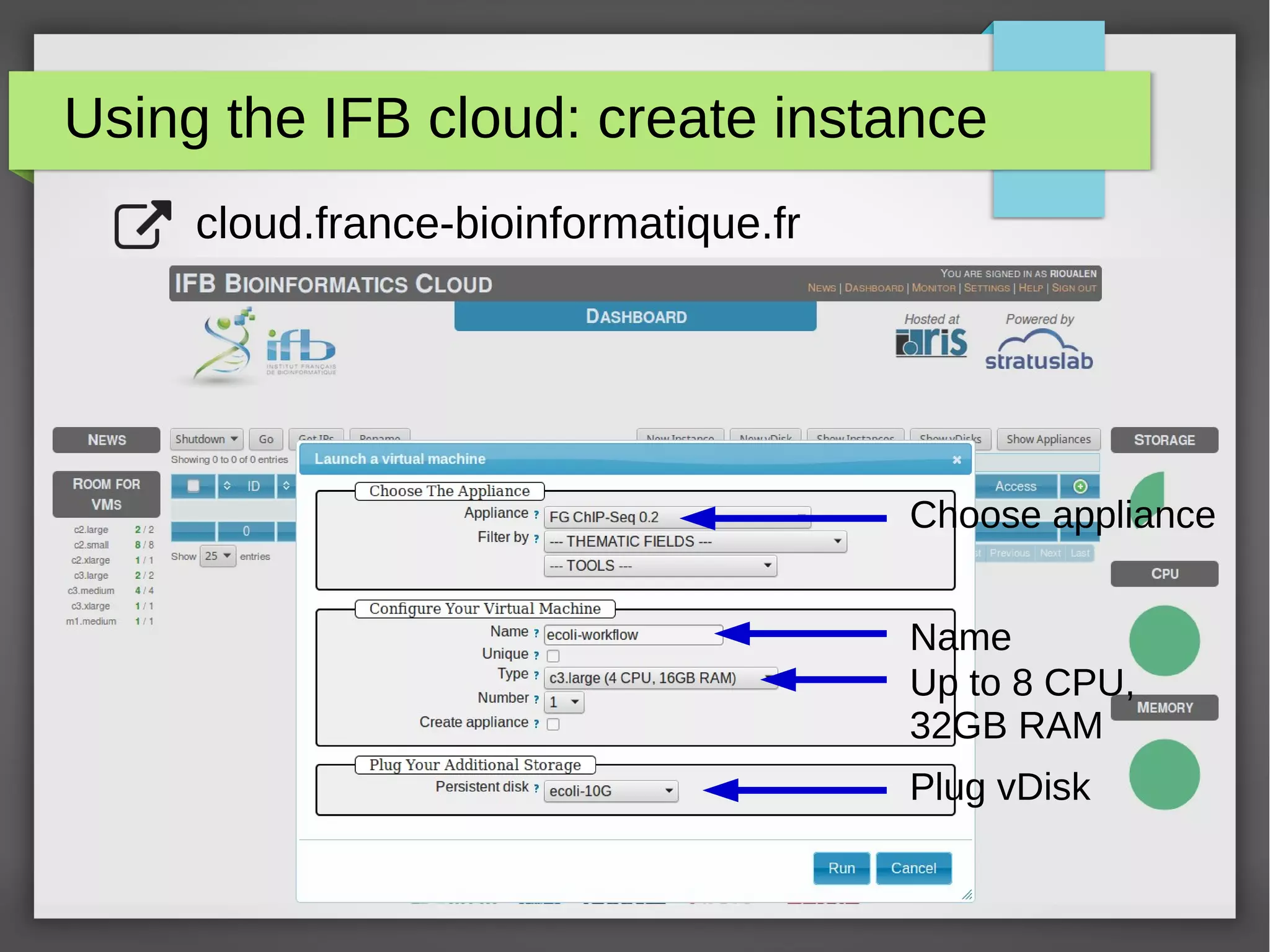
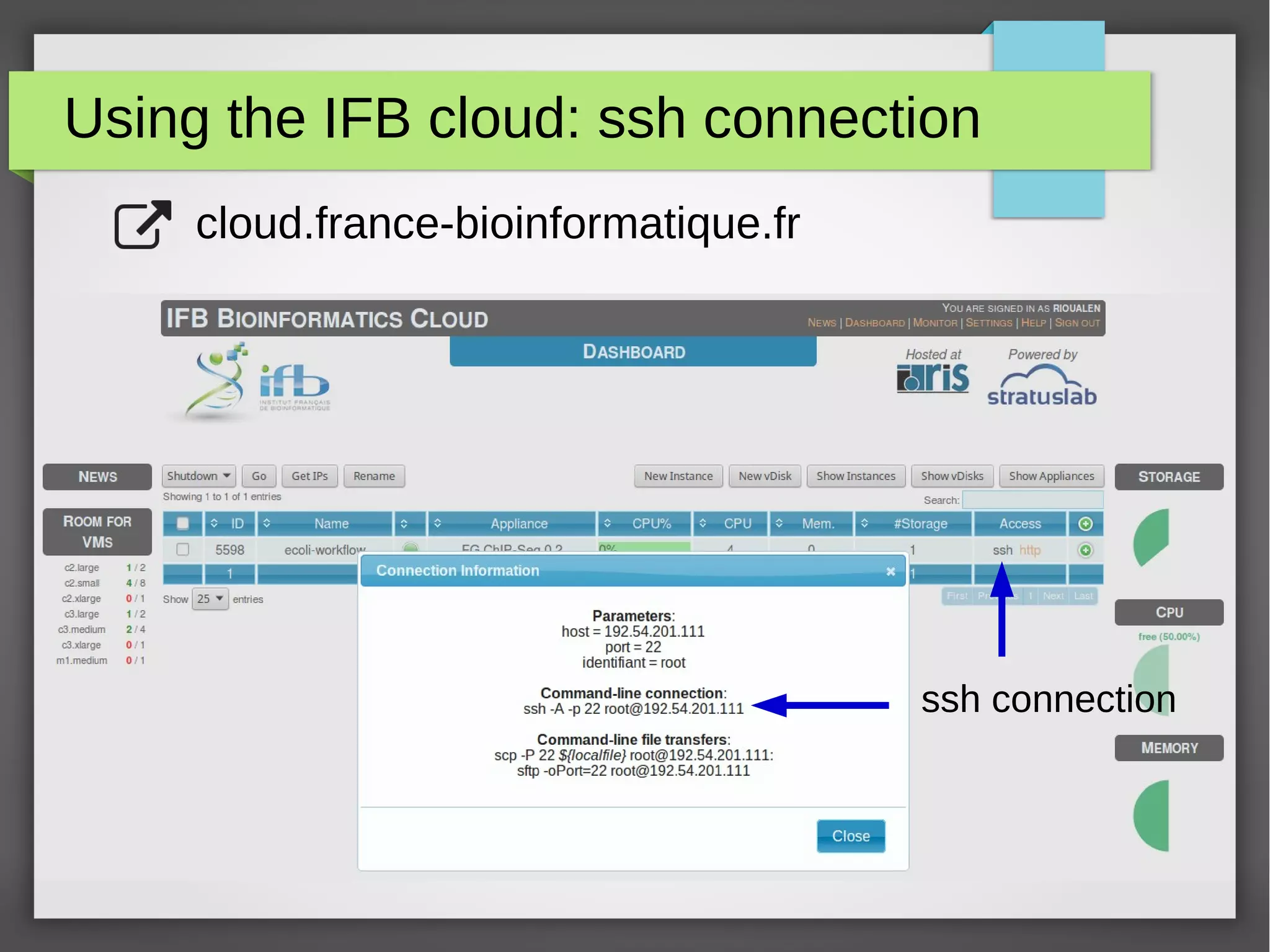
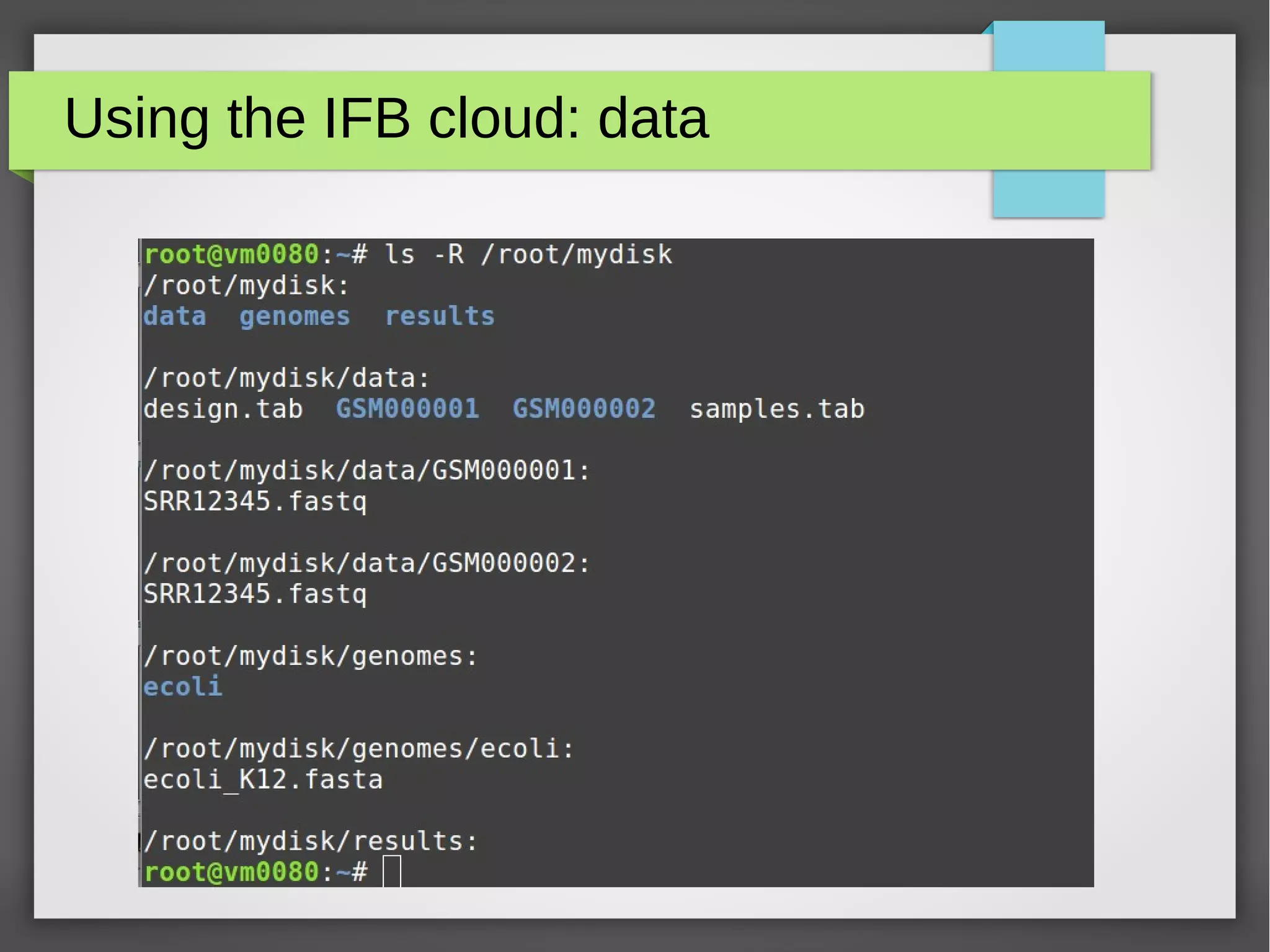
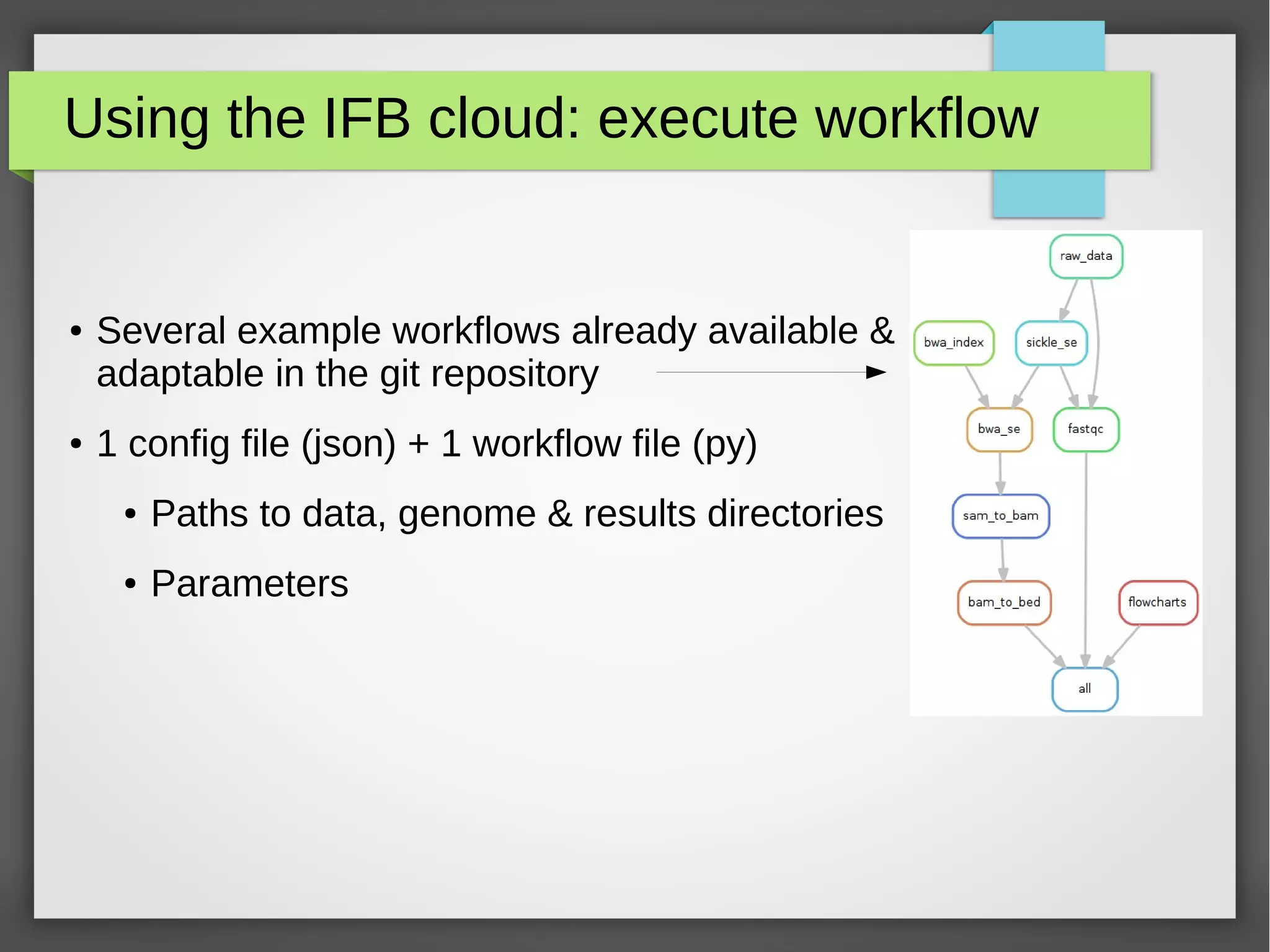
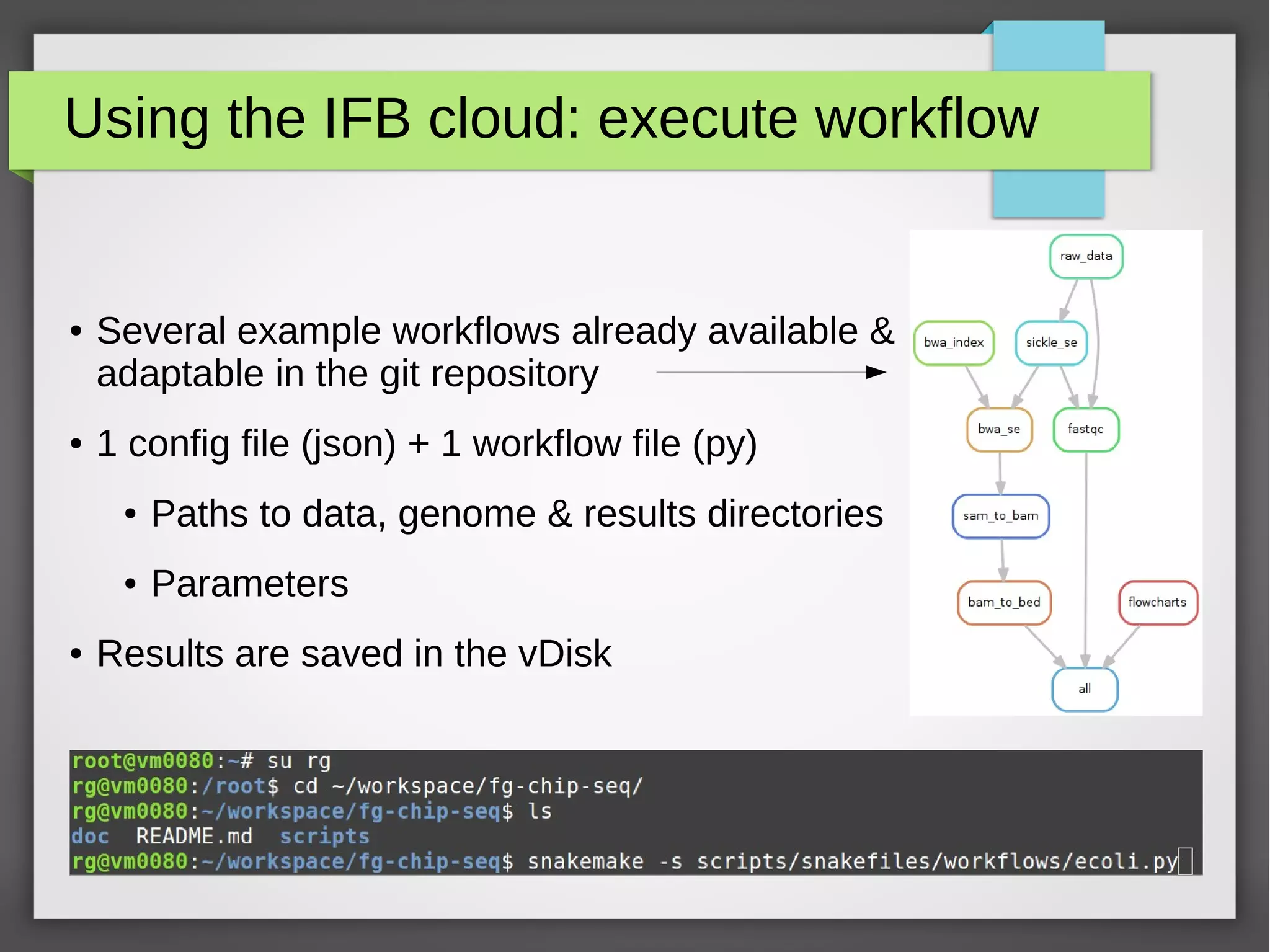
The document describes the IFB cloud, a French national bioinformatics infrastructure that provides computing resources and appliances. It summarizes the current and planned capacity of the IFB cloud. It then describes a ChIP-seq analysis appliance available on the IFB cloud, which contains tools like snakemake, NGS tools, and example ChIP-seq workflows. It provides instructions for requesting an account, launching a virtual disk and instance, connecting via SSH, loading data, and executing workflows on the appliance using the snakemake framework.