This document discusses using porphyrin-based nano-tiles for evolvable designs and computation through self-assembly. It outlines research on:
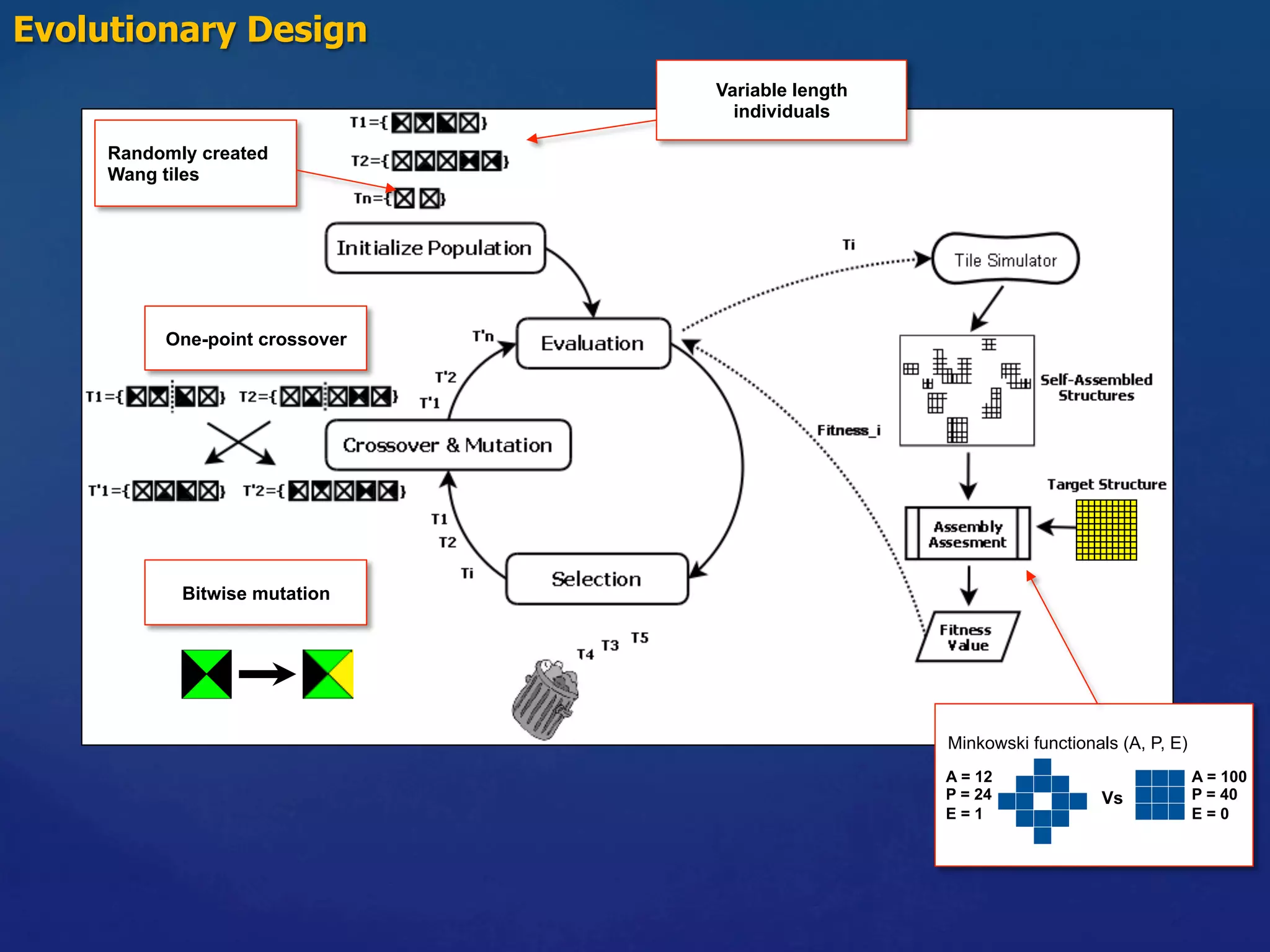
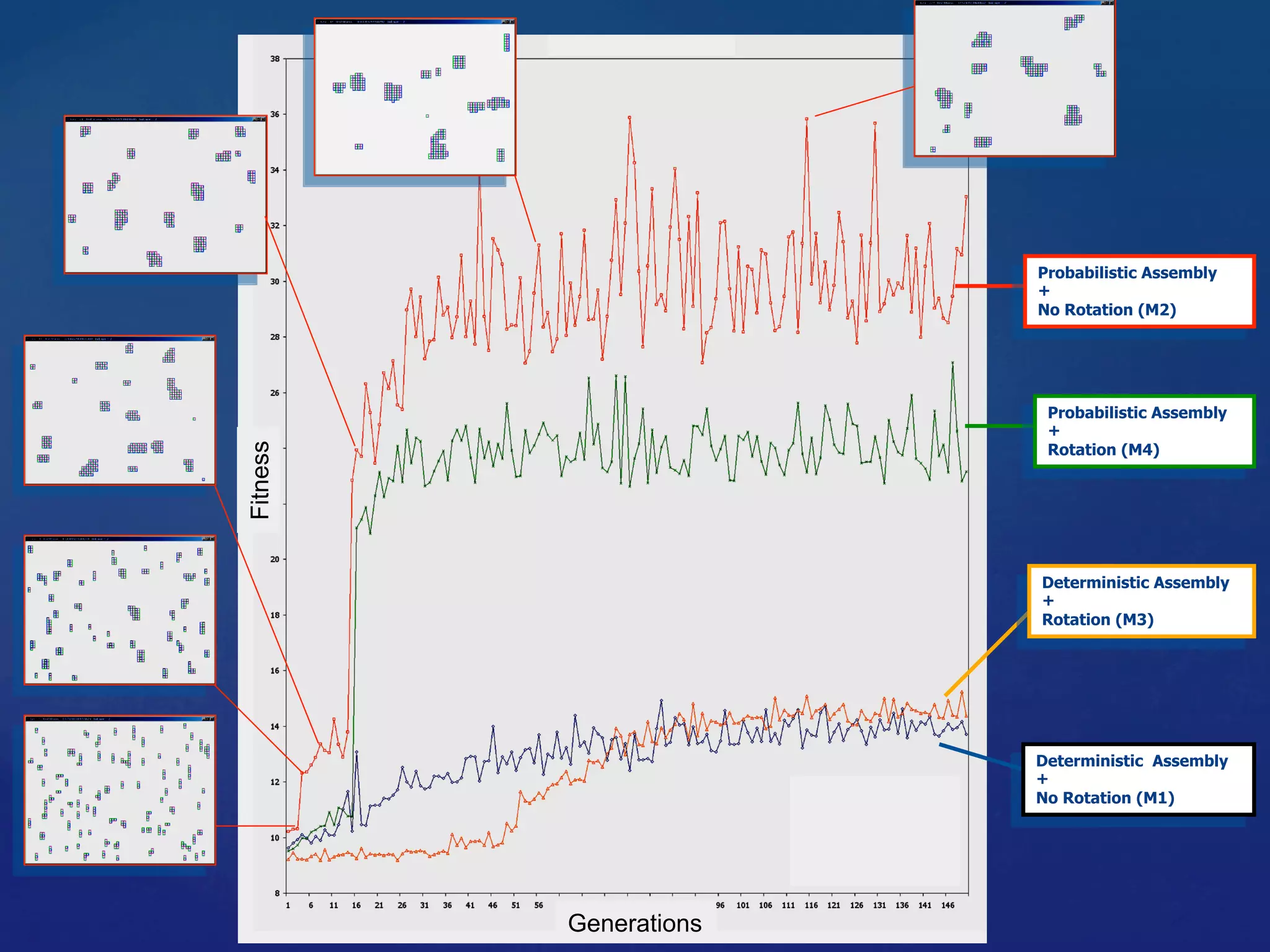
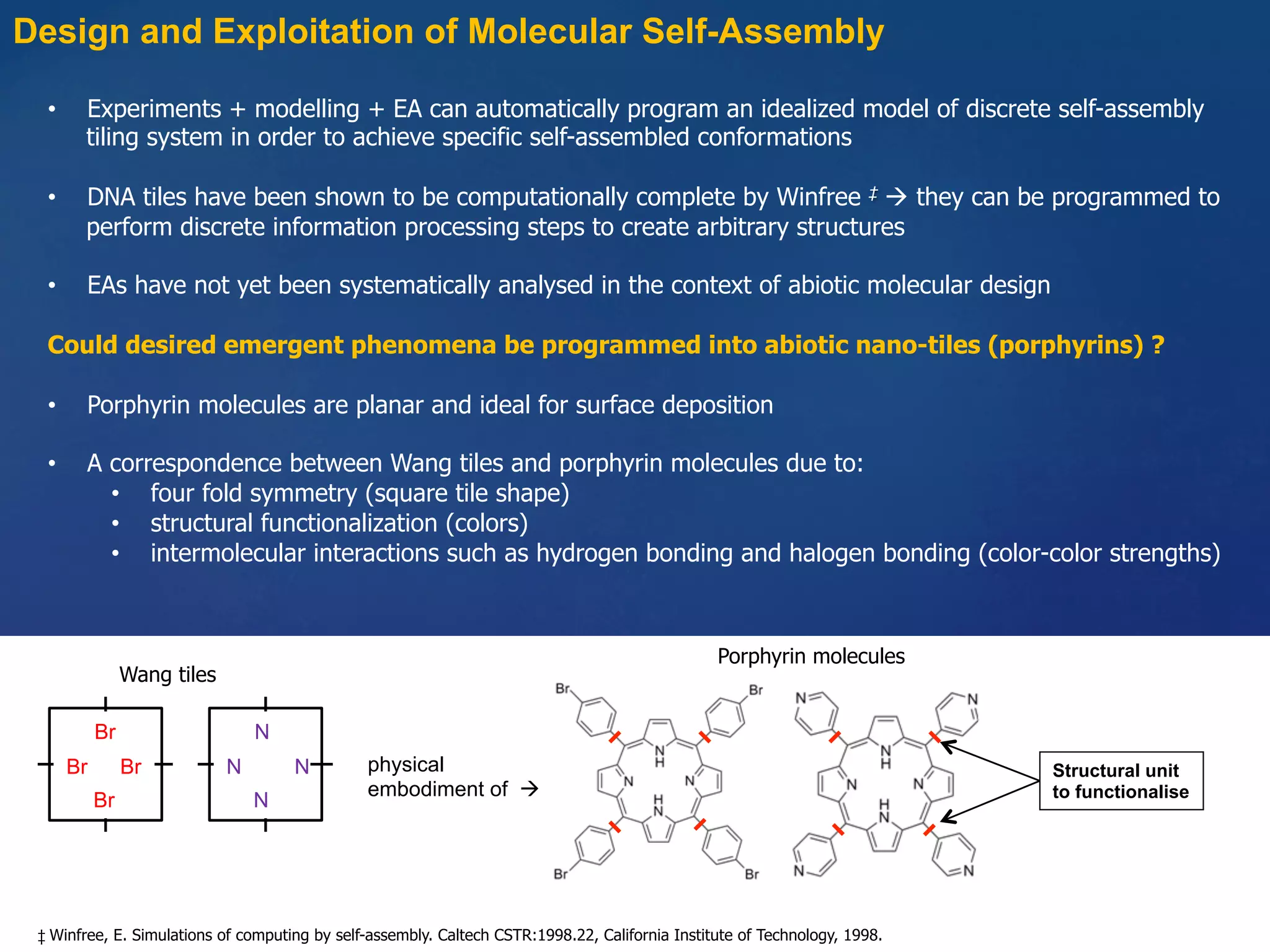
1) Using evolutionary algorithms to program self-assembly of Wang tiles to achieve target structures and applying this approach to programmable self-assembly of porphyrin molecules.
2) Modeling self-assembly of porphyrin molecules with different binding strengths using kinetic Monte Carlo simulations to predict experimental outcomes.
3) Analyzing self-assembled structures from the simulations using metrics like Minkowski functionals and Kolmogorov complexity to characterize computation and information processing during self-assembly.




![Self-assembly Wang Tiles Programmability Self-Assembly Wang Tiles
Target structures: square-like shapes
N tiles
Ntiles
Tiles System
• Finite size
square-site lattice
(300x300)
• Fixed T = 4
• Fixed M
Q: Is it possible to program
the family tiles needed to
obtain arbitrary structures by
means of SA ?
M [color, color] : strength
1 3 7 5 2
3 5 4 7 1
7 4 8 2 9
5 7 2 3 8
2 1 9 8 2](https://image.slidesharecdn.com/granada-160906112709/75/Evolvability-of-Designs-and-Computation-with-Porphyrins-based-Nano-tiles-7-2048.jpg)

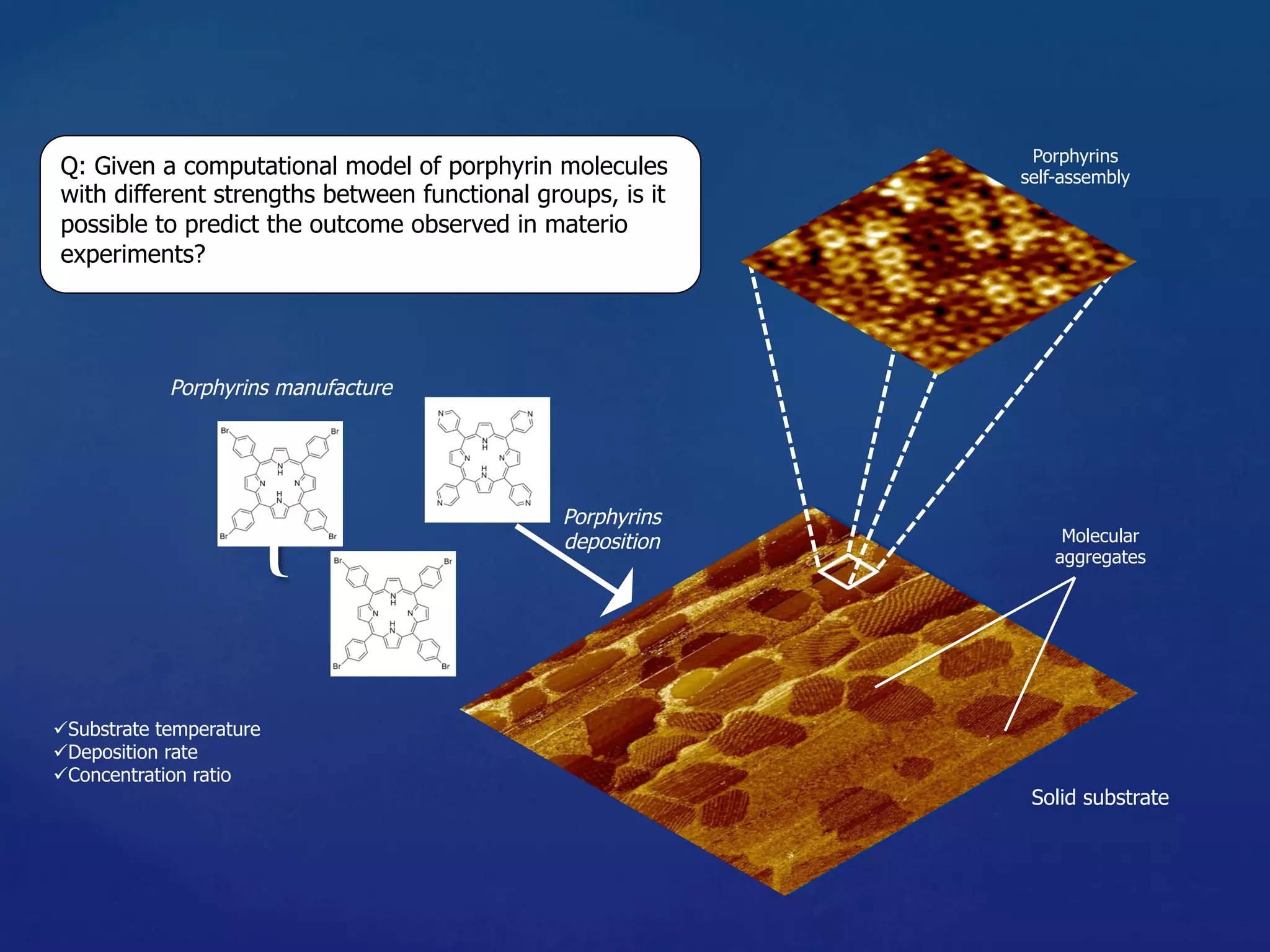
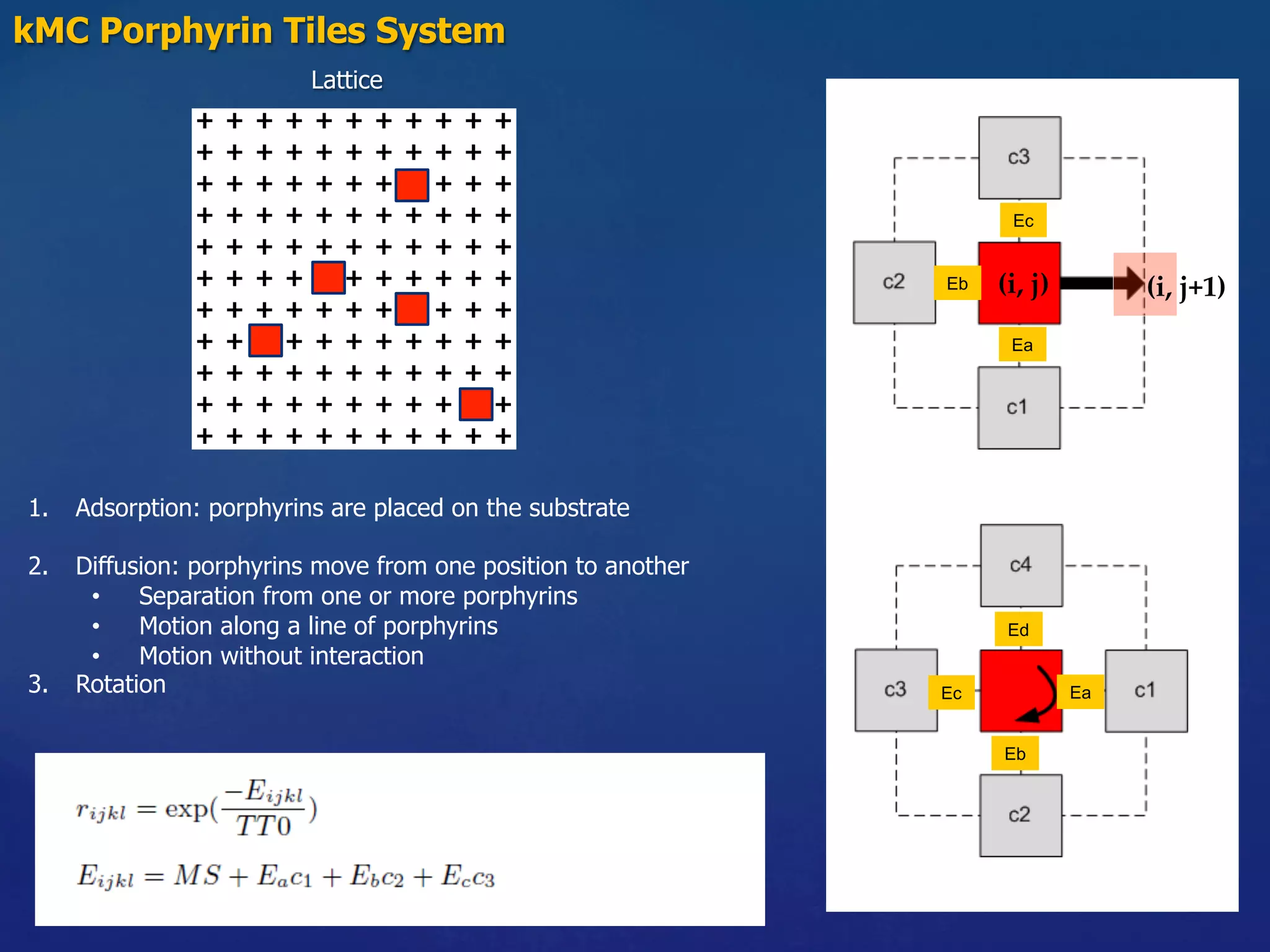
![Molecule types: m1, m2
Fixed Parameters
Substrate: 256 x 256
Coverage: 25%
Variable Parameters
Molecule-Substrate (MS) = [0.5, 1.0] res. 0.1 eV
Binding strength (BE11) = [0.1, 1.0] res. 0.1 eV
Binding strength (BE22) = [0.1, 1.0] res. 0.1 eV
Binding strength (BE12) = [0.1, 1.0] res. 0.1 eV
m1
1
1
1
1
2
m2 2
2
2
Experiments
m1 m1
m2 m2
m1 m2](https://image.slidesharecdn.com/granada-160906112709/75/Evolvability-of-Designs-and-Computation-with-Porphyrins-based-Nano-tiles-14-2048.jpg)
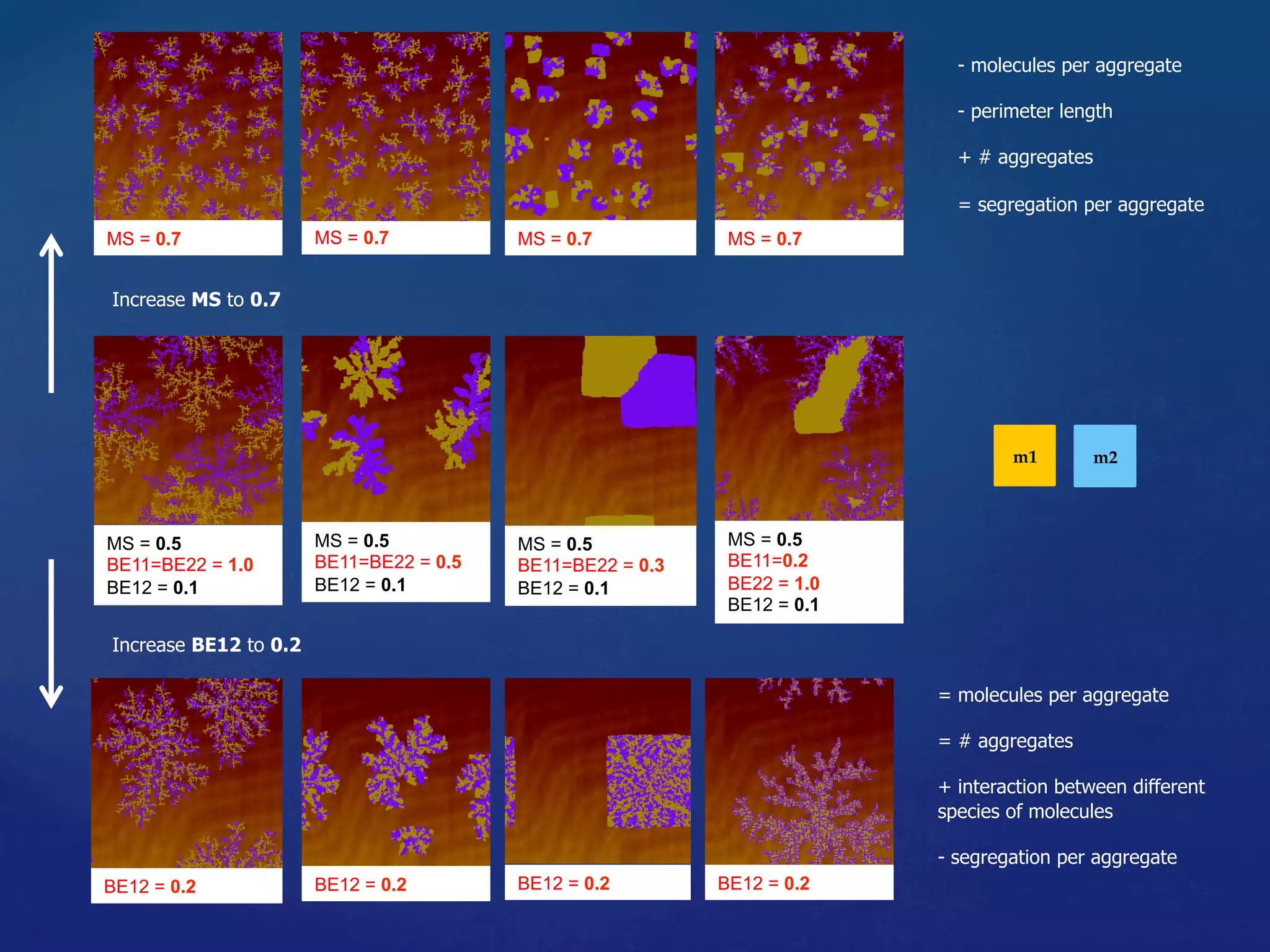

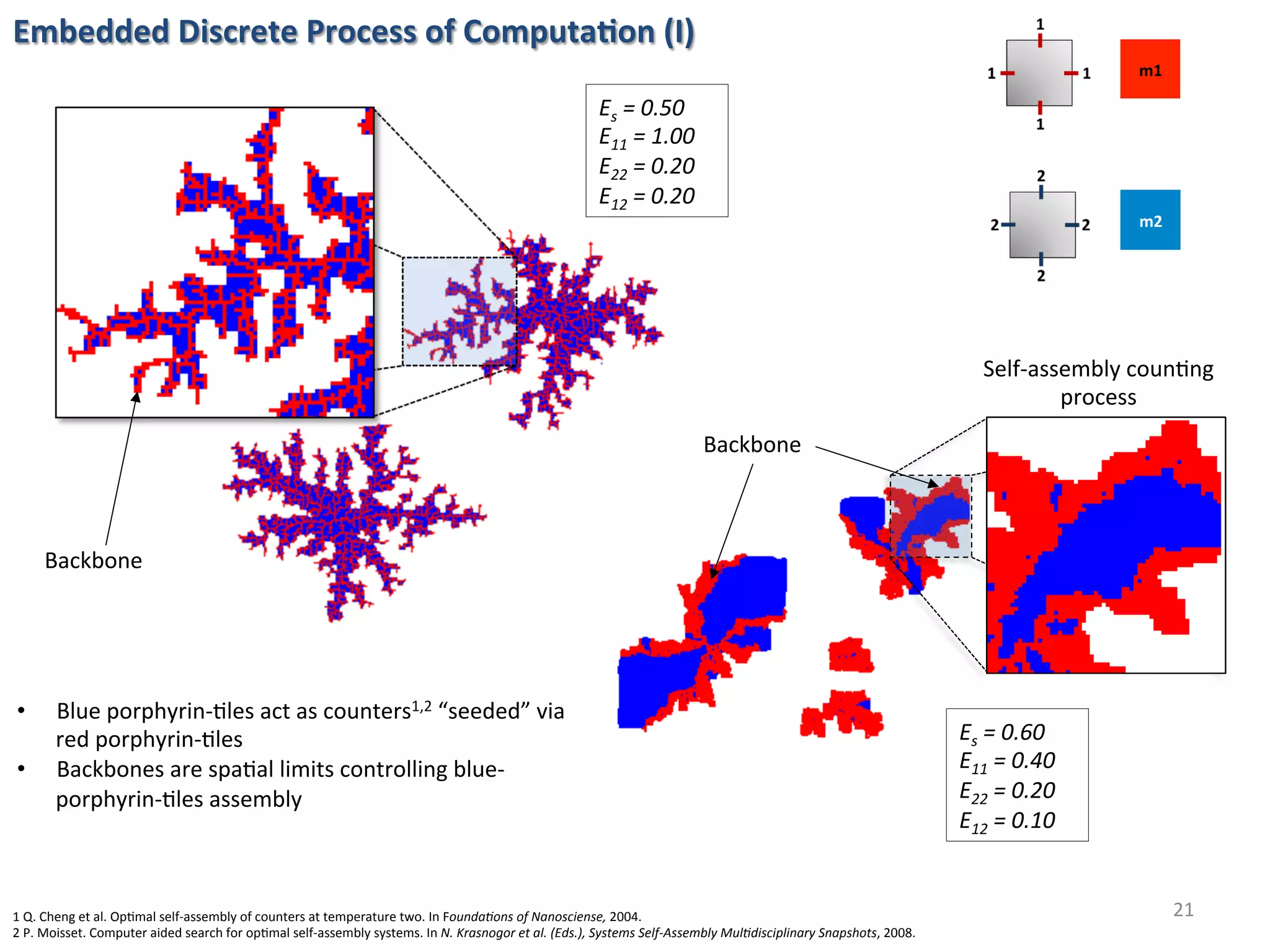
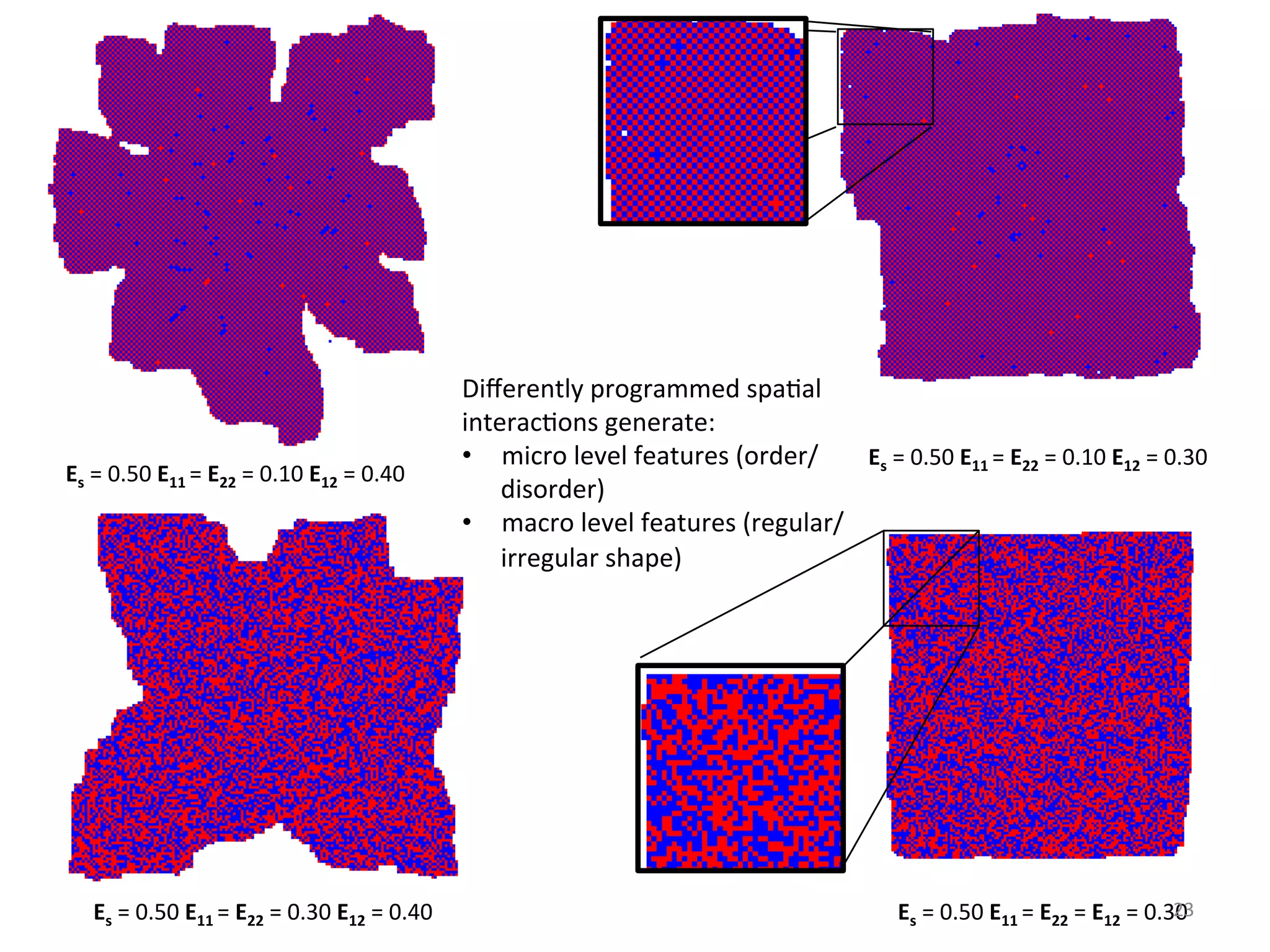
![Embedded Discrete Process of Computa%on (II)
22
Checkers paLern
(spa6al interac6ons)
• Highly ordered self-assembled structure
• Spontaneous internal arrangements
• Globally complex shape with locally
simple organisa6on
λ (y)
λ (y)
(x)
(ε) (ε)
(x)
q1
q2
ε, x, y Є [0, 1]
ε + x + y = 1
x >> ε >> y
Computed by a finite state
machine-like process
ε: probability of mistaking symbol
λ: new diagonal begins](https://image.slidesharecdn.com/granada-160906112709/75/Evolvability-of-Designs-and-Computation-with-Porphyrins-based-Nano-tiles-22-2048.jpg)
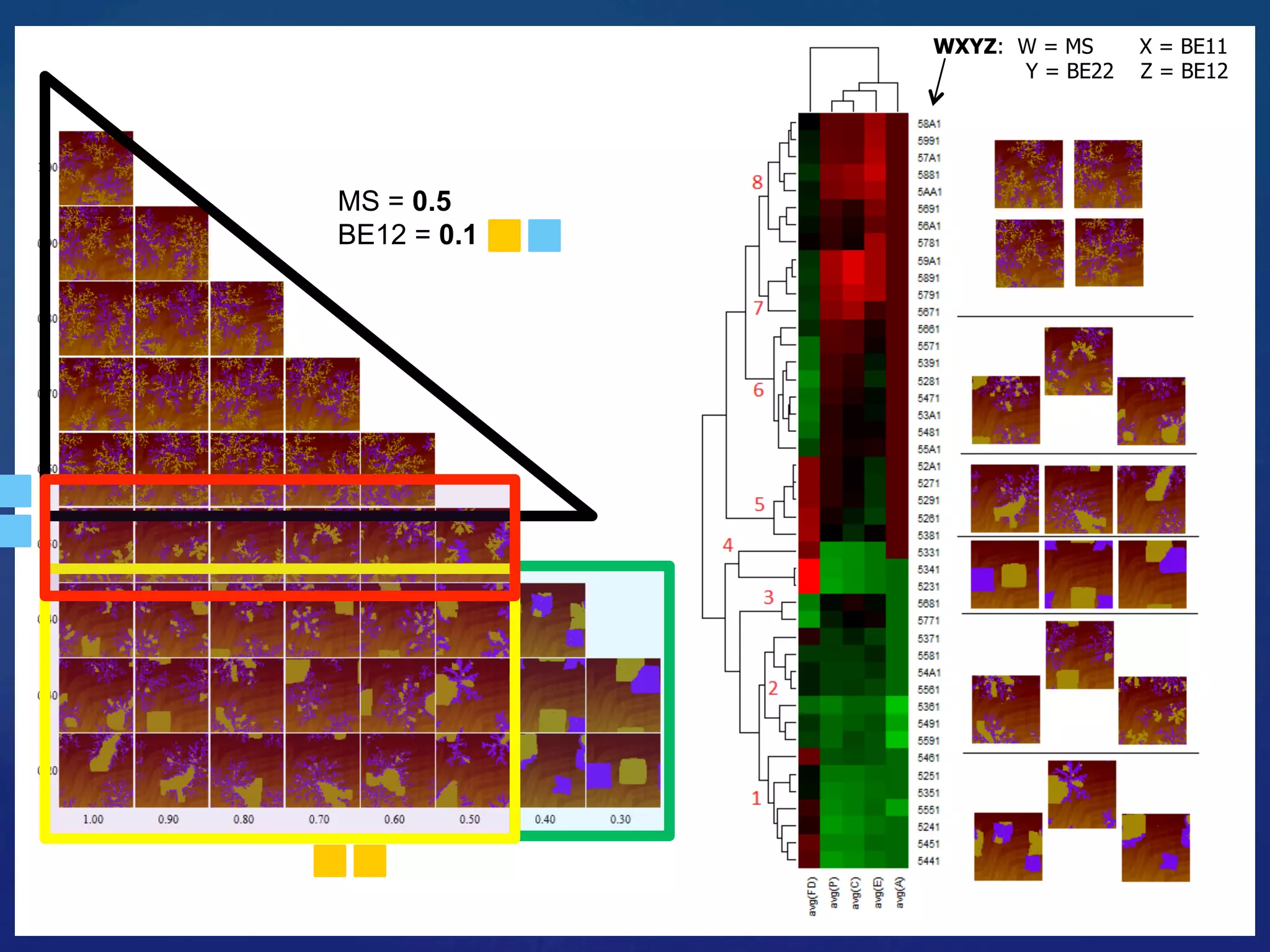
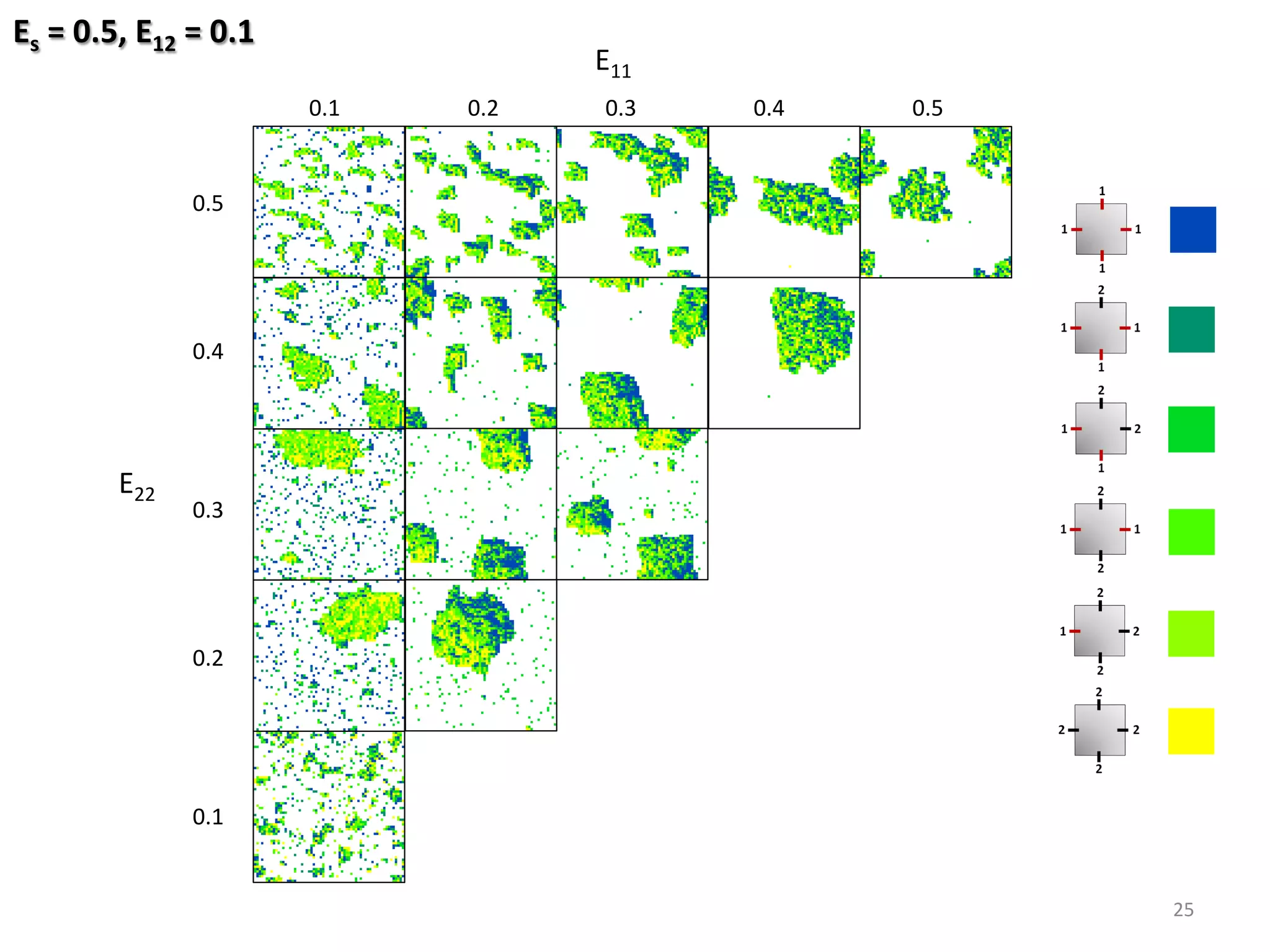
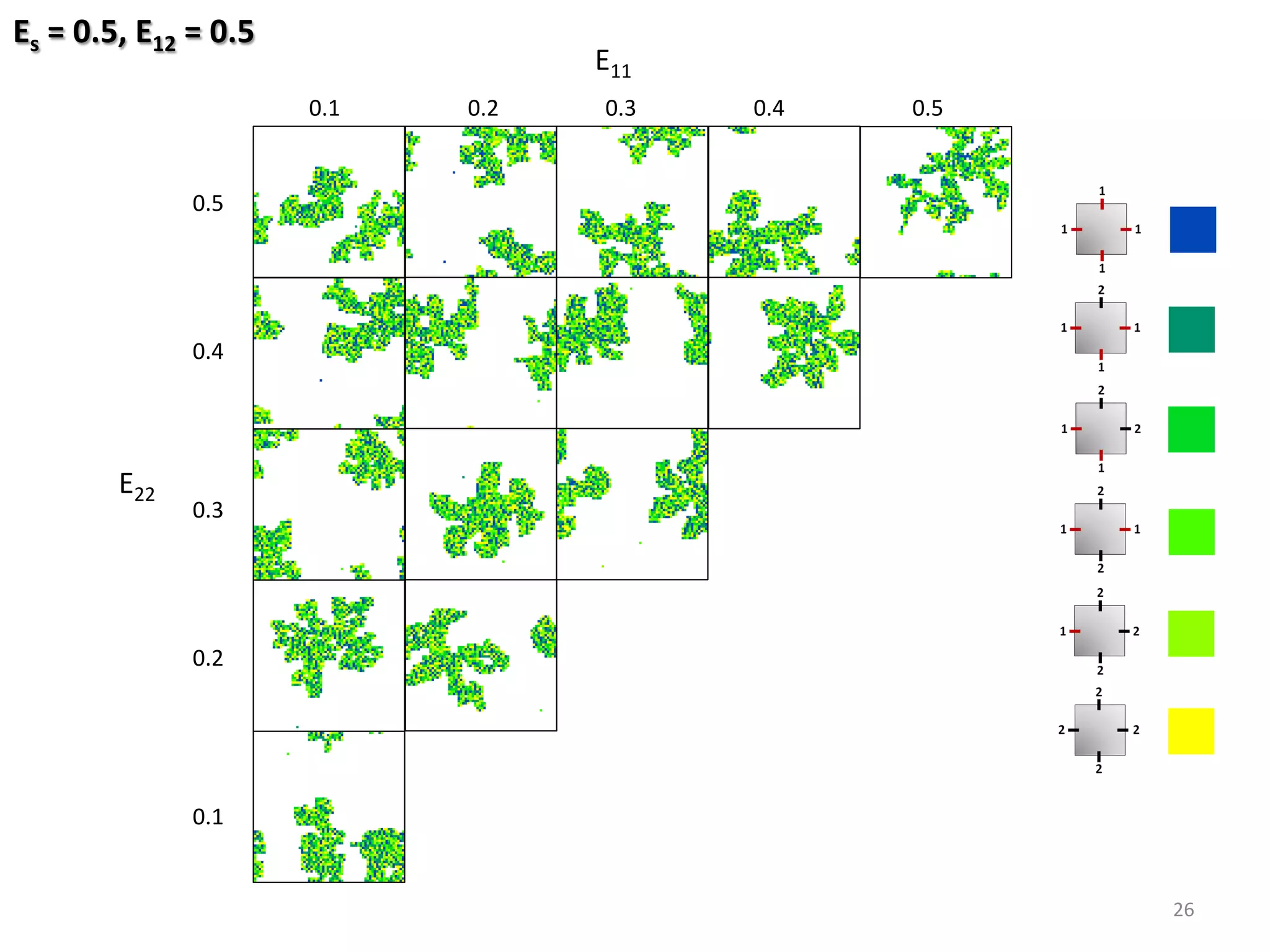
![Fixed Parameters
Substrate: 64 x 64
Coverage: 25%
Variable Parameters
Molecule-Substrate (ES) = [0.5, …, 0.7] res. 0.1 eV
Binding strength (E11, E22) = [0.1, …, 0.5] res. 0.025 eV
Binding strength (E12) = [0.1, …, 0.5] res. 0.1 eV
24
Hetero-func/onalised Porphyrin-%les Species:](https://image.slidesharecdn.com/granada-160906112709/75/Evolvability-of-Designs-and-Computation-with-Porphyrins-based-Nano-tiles-24-2048.jpg)
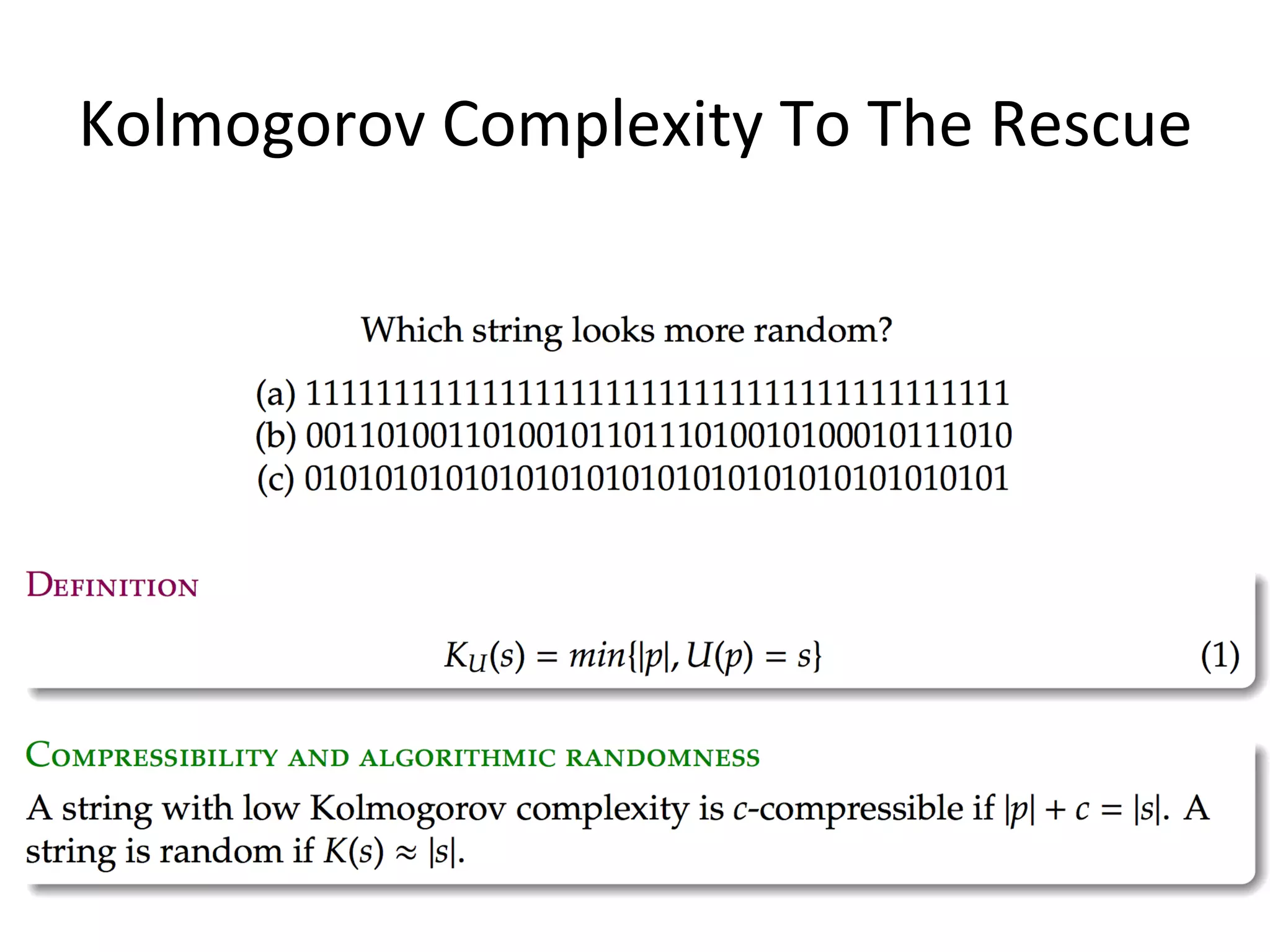

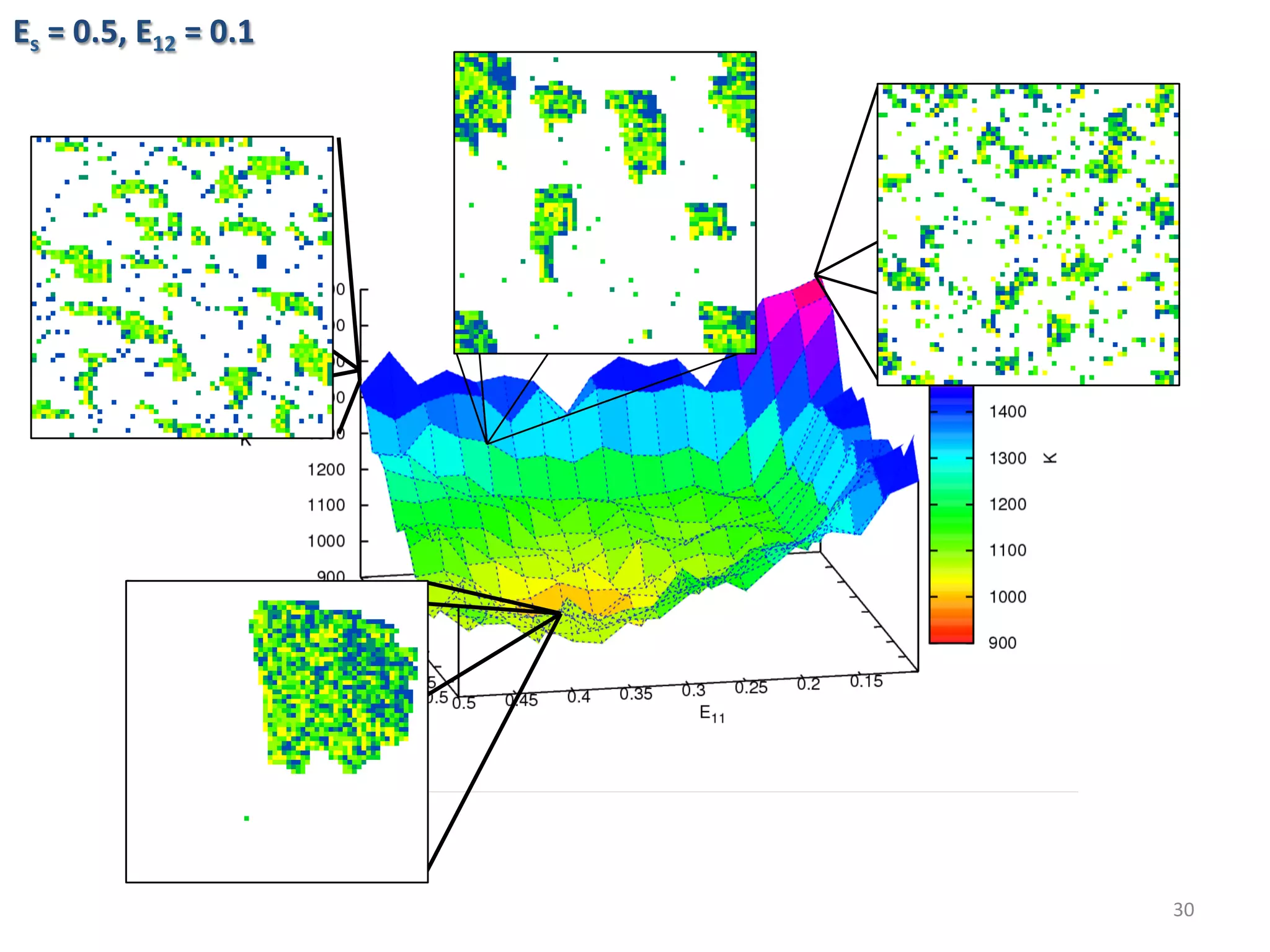
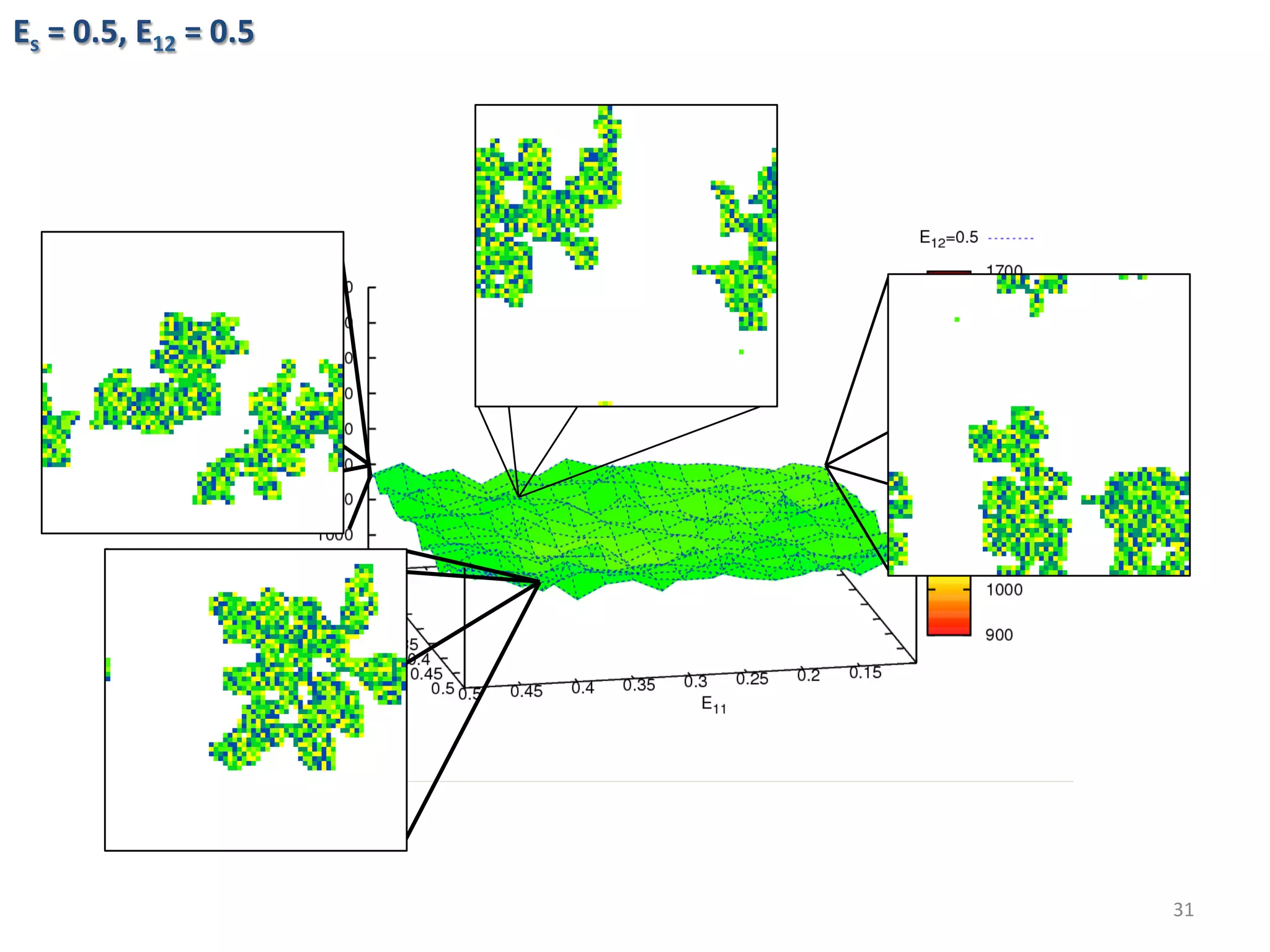
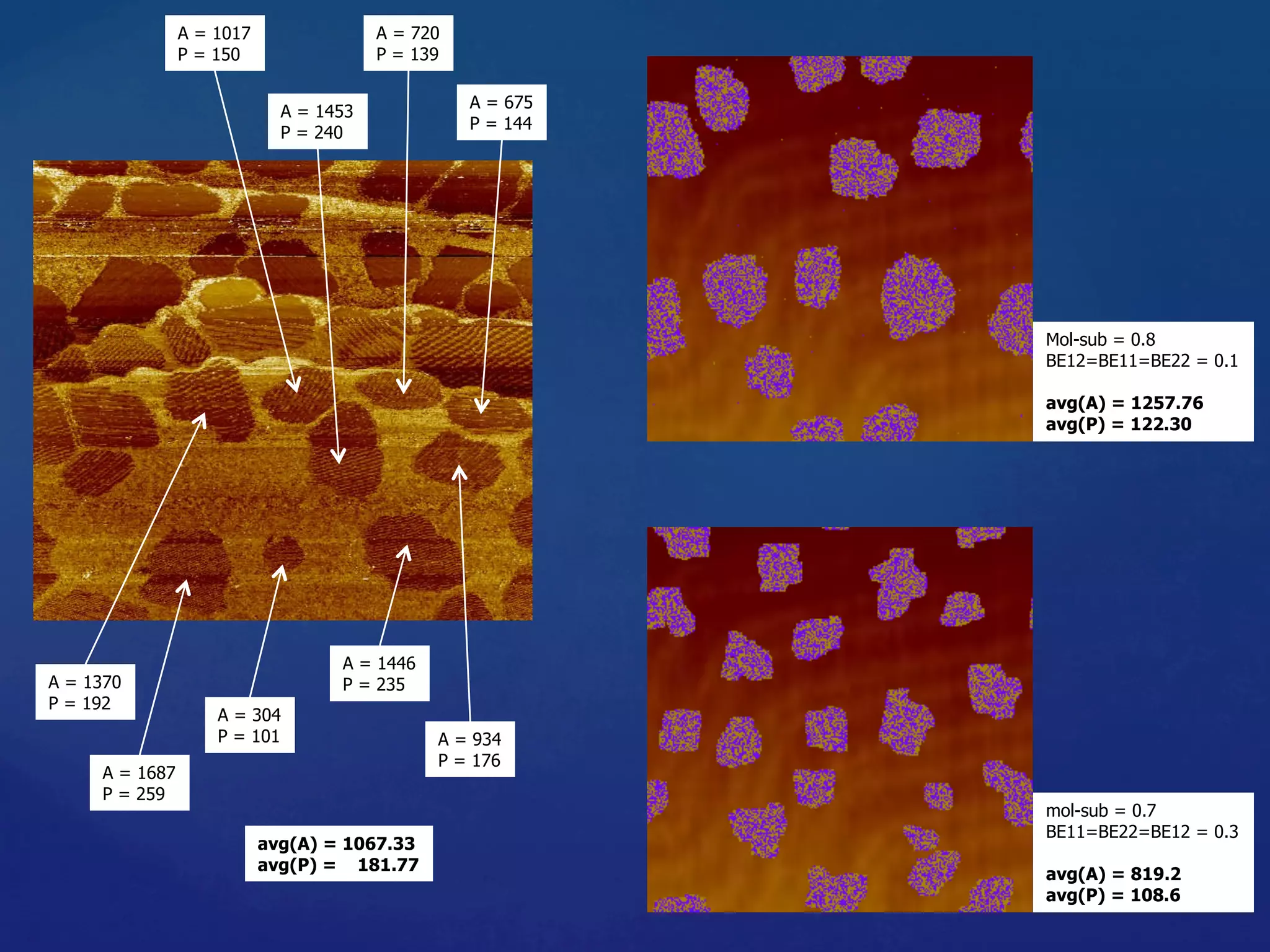
![Algorithmic Informa6on Content
• Algorithmic Complexity of a string s, K(s),
The length of the shortest program, p, that could generate
the string.
• K is an uncomputable func6on. A prac6cal way to
approximate K is using lossless compression algorithms.
• The outputs of the simula6ons are converted into PNG
images then compressed using PNGcrush. The compressed
size of the images are the es6mated algorithmic complexity
of the outputs.
29
)})(],[Length{min()( spUpsK ==](https://image.slidesharecdn.com/granada-160906112709/75/Evolvability-of-Designs-and-Computation-with-Porphyrins-based-Nano-tiles-29-2048.jpg)