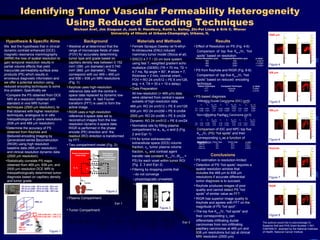
The document investigates the hypothesis that spatial resolution loss in dynamic contrast-enhanced magnetic resonance mammography leads to inaccuracies in tumor vascular permeability assessments. It compares permeability-surface area products obtained using reduced encoding techniques at different resolutions to determine their accuracy in differential tumor diagnosis. The study concludes that higher spatial resolutions are necessary for accurate identification of tumor types, with reduced encoding techniques like RIGR performing better than keyhole imaging.