This document discusses biohackathons, which are events where participants collaborate to create new tools or applications in bioscience over a weekend. It provides examples of two biohackathons held in 2014 at Stanford University and University of California San Diego. At these events, around 30 participants formed teams and worked on projects like tools for visualizing and editing bio data or finding similarities between biomedical concepts. The goals were to foster collaborative creativity, learning, and networking. The best projects received small cash prizes.









![Projects
• BioPolymer: A set of embeddable web components to display and edit bio data. Initially data from MyGene &
MyVariant. Team: Mark Fortner, Keiichiro Ono
• SameSame: a dynamic tool for finding and visualizing the degree of similarity between any set of biomedical
concepts. Team: Benjamin Good, Maulik Kamdar, Alan Higgins, Alex Williams
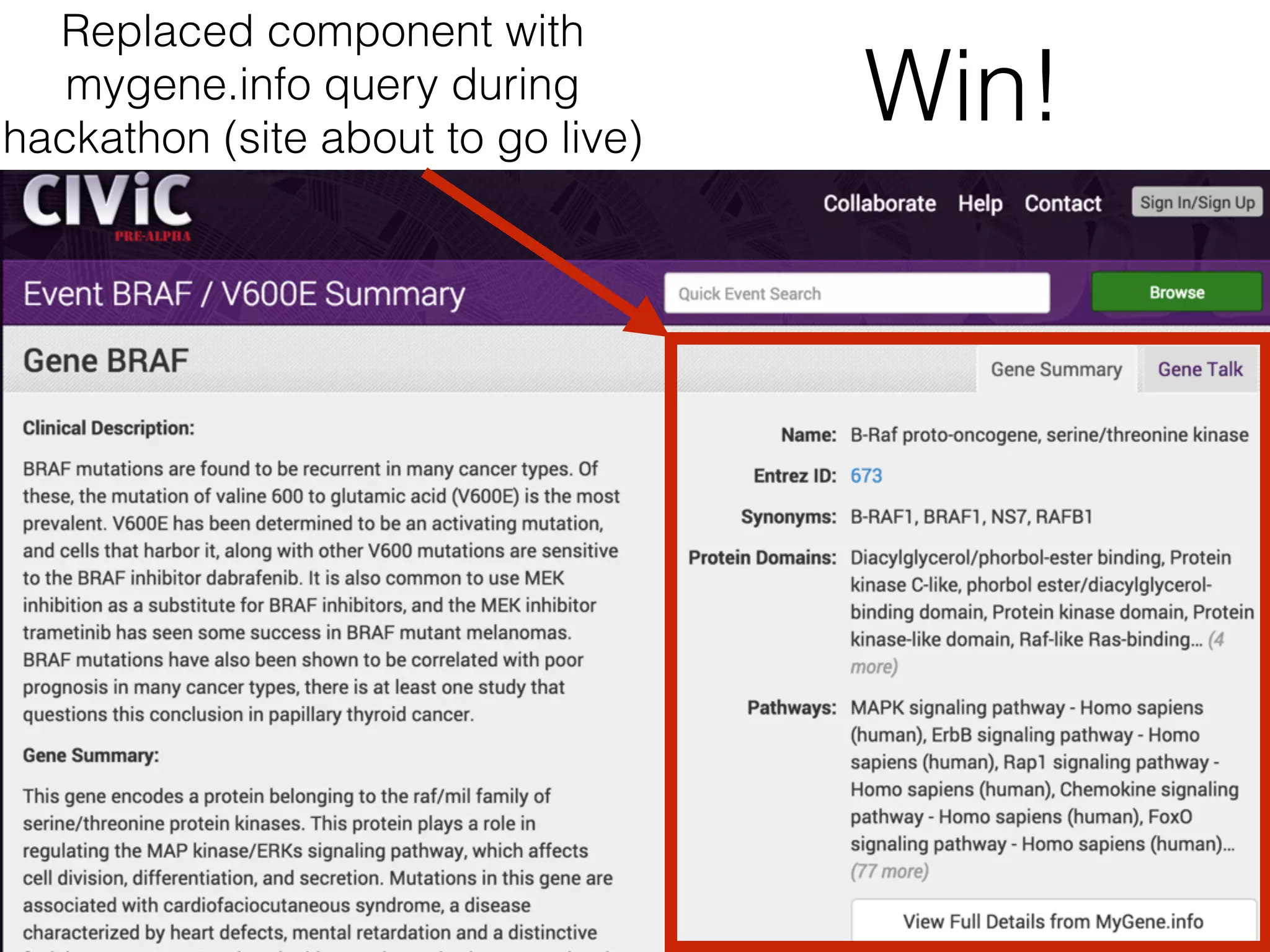
• CIViC - Clinical Interpretation of Variants in Cancer: Crowdsourcing and web interface for curation of clinically
actionable evidence in cancer. Team: Obi Griffith, Adam Coffman, Martin Jones, Karthik G, Jon Cristensen, Julianne
Schneider
• Citizen Science: An app to enable people to extract structured ‘facts’ (subject predicate object triples) from
unstructured text. Here is the project presentation. Team: Richard Good, Hannes Niedner, Andrew Su
• SBiDer: Synthetic Biocircuit Developer: A web-app to search a database of operons [functional biochemical
pathways] to use them in new and novel ways [to make synthetic organisms such as the ones used to make this
Malaria treatment]. Team Justin Huang, Kwat Yeerna, Fernando Contreras, Joaquin Reyna, Jenhan Tao
• NDex: The NDEx project provides a public website where scientists and organizations can share, store, manipulate,
and publish biological network knowledge. Team Dexter Pratt
• fiSSEA: A framework that integrates MyGene.info and MyVariant.info to retrieve functional prediction annotations (or
any type of annotation) for knowledge discovery, specifically implement CADD scores for “functional impact SNP Set
Enrichment Analysis". Team: Adam Mark, Erick Scott, Chunlei Wu
• MyGene.Info Taxonomy Query: Added detailed taxonomy information to mygene.info. Allows queries based on
taxonomy ID and advanced queries based on hierarchies of taxonomic nodes. Team: Greg Stupp, Chunlei Wu](https://image.slidesharecdn.com/hackathonbd2k2015pdf-150212021603-conversion-gate01/75/Bio-Hackathons-11-2048.jpg)