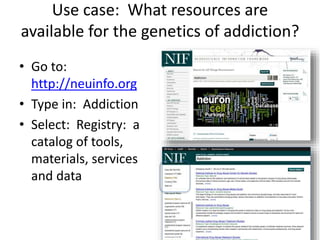
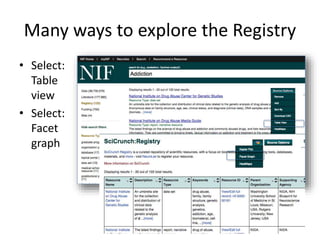
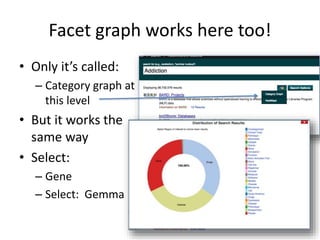
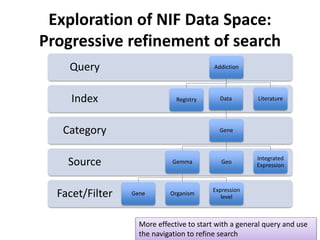
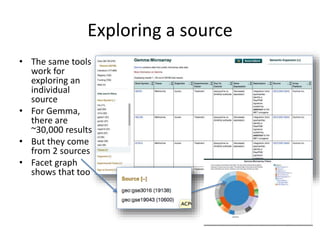
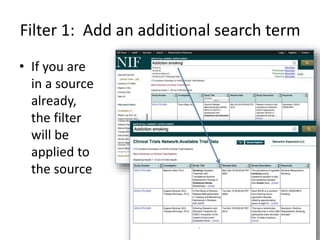
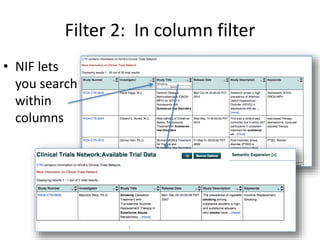
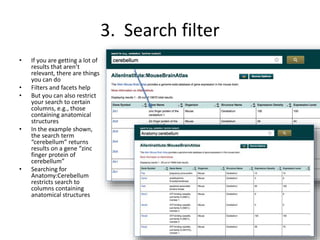
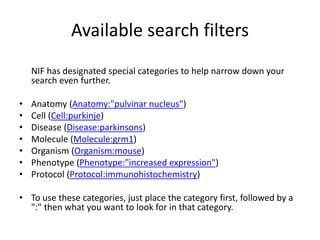
This document outlines a guided tutorial on the neuroscience information framework (NIF) registry and data federation for a workshop on genetics of addiction. It details how to find and explore various resources and databases within the NIF, including methods for searching, filtering, and refining queries related to addiction research. Additionally, it provides instructions on adding resources to the NIF registry and emphasizes the importance of effective searching techniques to manage extraneous results.