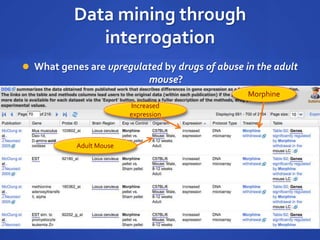
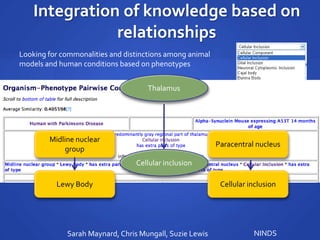
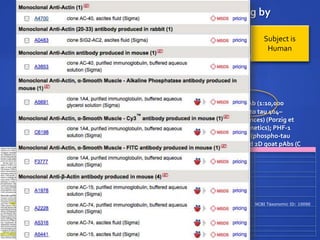
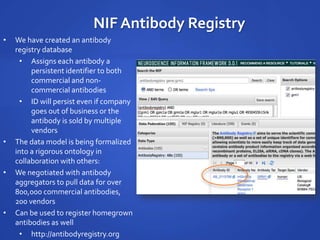
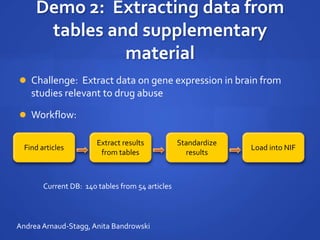
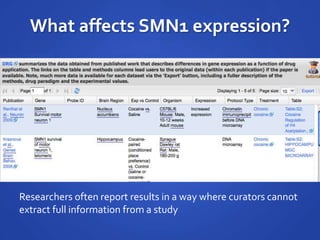
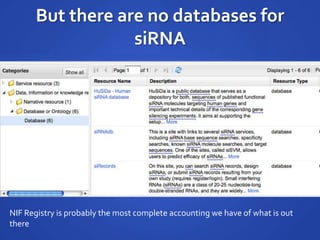
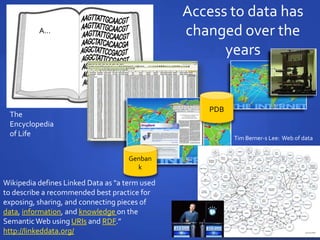
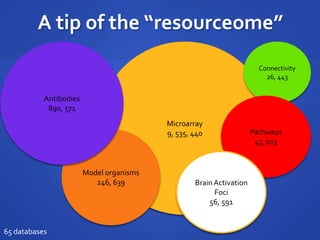
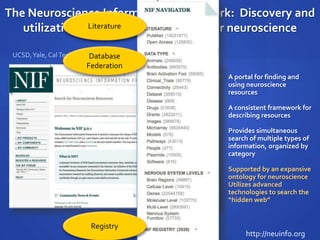
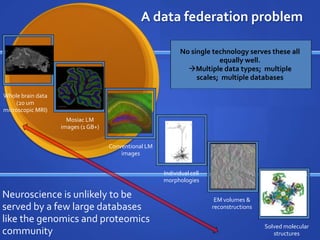
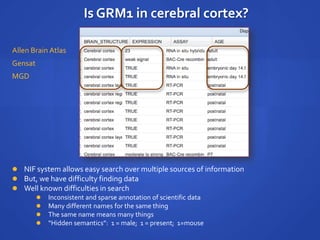
The document discusses the challenges of establishing a global neuroscience information framework, emphasizing the need for improved data integration, standardization, and accessibility within the neuroscience community. It highlights the inadequacies of current data storage practices, the lack of common metadata, and various barriers to effectively utilizing available data. Proposed solutions include developing analytical tools, robust ontologies for data representation, and enhancing collaborative efforts to better understand brain functions through organized and linked data.















![Linking datatypes to semantics: What is
the average diameter of a Purkinje
neuron dendrite?
Branch structure not a
tree, not a set of blood
vessels, not a road map but a
DENDRITE
Because anyone who uses
Neurolucida uses the same
concepts: axon, dendrite, cell
body, dendritic
spine, information systems
can combine the data
together in meaningful ways
Neurolucida
doesn’t, however, tell you that
dendrite belongs to a neuron
of a particular type or whether
this dendrite is a neural
dendrite at all
( (Color Yellow) ; [10,1]
(Dendrite)
( 5.04 -44.40 -89.00 1.32) ; Root
( 3.39 -44.40 -89.00 1.32) ; R, 1
(
( 2.81 -45.10 -90.00 0.91) ; R-1, 1
( 2.81 -45.18 -90.00 0.91) ; R-1, 2
( 1.90 -46.01 -90.00 0.91) ; R-1, 3
( 1.82 -46.09 -90.00 0.91) ; R-1, 4
( 0.91 -46.59 -90.00 0.91) ; R-1, 5
( 0.41 -46.83 -92.50 0.91) ; R-1, 6
(
( -0.66 -46.92 -88.50 0.74) ; R-1-1, 1
( -0.74 -46.92 -88.50 0.74) ; R-1-1, 2
( -2.15 -47.25 -88.00 0.74) ; R-1-1, 3
( -2.15 -47.33 -88.00 0.74) ; R-1-1, 4
( -3.06 -47.00 -87.00 0.74) ; R-1-1, 5
( -4.05 -46.92 -86.00 0.74) ; R-1-1, 6
Output of Neurolucida neuron trace](https://image.slidesharecdn.com/2010-martone-globalinformationframe-130612165114-phpapp01/85/The-possibility-and-probability-of-a-global-Neuroscience-Information-Framework-23-320.jpg)