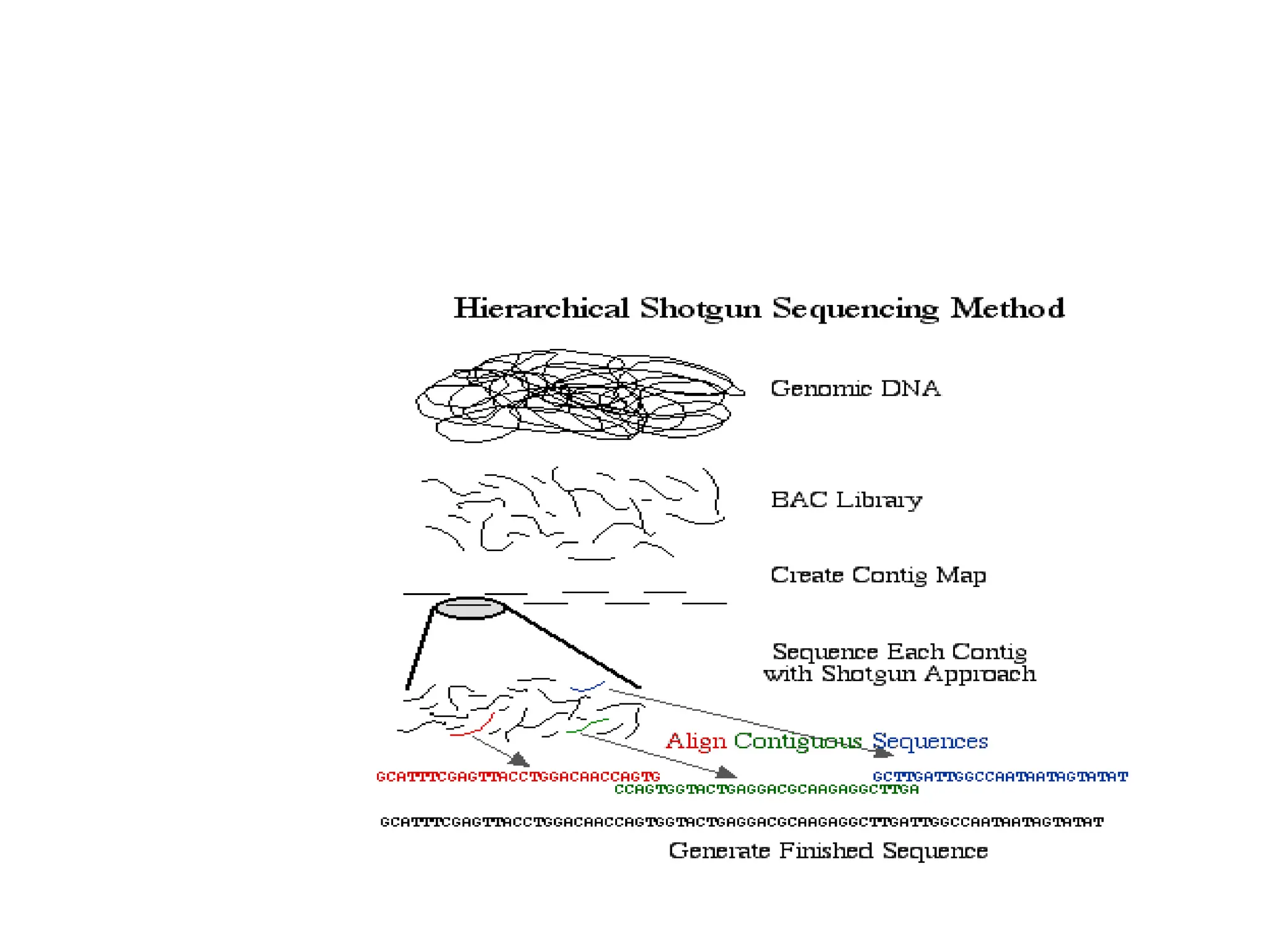
The document discusses genome sequencing, detailing its definition, methods, and steps in executing it. It focuses on hierarchical shotgun sequencing, preferred by the Human Genome Project, and whole genome shotgun sequencing developed by Craig Venter and H. Smith, describing their distinct processes. Additionally, it introduces hybrid shotgun sequencing, which combines elements of both methods for efficient genome analysis.