This document proposes collaborating with the BioDBCore initiative to standardize the registration and description of biological databases. It identifies challenges in uniquely identifying databases due to unstable URLs. The proposal suggests adopting the MIRIAM registry's persistent identifiers to decouple identification from location. Benefits include globally identifying life science databases, improved discovery of relevant resources, and potential for BioDBCore to evolve into a database publishing platform. Open questions remain regarding technical details and integrating existing database lists.



![G. A. Thorisson, A. J. Webb, R. Dalgleish ULEIC / J. Muilu FIMM
How might this work?
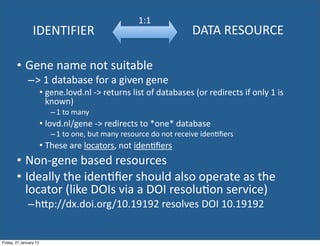
• Using database URIs - plausible scenario
– Persistent canonical URI: http://identifiers.org/biodbcore/10235900
– Click URL, browser redirects to http://biodbcore.org/resource/10235900
– BioDBCore metadata record for the database (akin to “landing page” online journal
site)
• BioDBCore “landing page” presents database metadata
– Information *about* the “thing”
– Name: Ehlers-Danlos Syndrome Variant Database
Main resource URL: https://eds.gene.le.ac.uk <-- the “thing” itself
[scope, data standards, other metadata]
• Location of database = the “thing” itself
GEN2PHEN 8th General Assembly Meeting, Leiden, Jan 24-25 2012 8
Friday, 27 January 12](https://image.slidesharecdn.com/gen2phengam8leidenidentifiersforlsdbsjan2012-120127045222-phpapp02/85/GEN2PHEN-GAM8-meeting-Leiden-Identifiers-for-LSDBs-8-320.jpg)